








生物技术通报 ›› 2022, Vol. 38 ›› Issue (4): 193-201.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1065
赵曾强1,2( ), 郭文婷1, 张析1, 李潇玲1, 张薇1(
), 郭文婷1, 张析1, 李潇玲1, 张薇1( )
)
收稿日期:2021-08-19
出版日期:2022-04-26
发布日期:2022-05-06
通讯作者:
张薇,女,博士,教授,研究方向:棉花分子育种;E-mail: zhw_agr@shzu.edu.cn作者简介:赵曾强,男,硕士,助理研究员,研究方向:棉花分子育种;E-mail: tlx4109@126.com
基金资助:
ZHAO Zeng-qiang1,2( ), GUO Wen-ting1, ZHANG Xi1, LI Xiao-ling1, ZHANG Wei1(
), GUO Wen-ting1, ZHANG Xi1, LI Xiao-ling1, ZHANG Wei1( )
)
Received:2021-08-19
Published:2022-04-26
Online:2022-05-06
摘要:
研究陆地棉ERF(ethylene-responsive factor)转录因子基因GhERF5-4D在棉花抗枯萎病中的作用,为棉花抗病分子机制及棉花抗病育种研究奠定基础。以枯萎病菌诱导棉花根部基因表达谱中的差异表达基因为探针,结合棉花全基因组数据库获得1个ERF转录因子基因GhERF5-4D,并进一步利用实时荧光定量(qRT-PCR)技术和病毒诱导基因沉默(virus induced gene silencing,VIGS)技术研究枯萎病菌胁迫和激素处理后基因GhERF5-4D的表达特征及其在棉花抗病过程中的作用。结果表明,GhERF5-4D(GenBank:MF093148)位于陆地棉D07染色体上,开放阅读框(open reading frame,ORF)为663 bp,编码220个氨基酸,仅含1个AP2结构域且无内含子,属于ERF亚族。该基因的表达特征在棉花抗感枯萎病品种及不同激素胁迫处理中存在显著差异,其在抗病品种中为上调表达,在感病品种中为下调表达;乙烯、茉莉酸甲酯处理后,该基因上调表达,而水杨酸处理后则下调表达。GhERF5-4D基因沉默后并进行枯萎病菌接种实验,结果表明,与对照相比,沉默株系更感病。对下游抗病相关防卫基因表达特征进行分析,发现PR2、PR4、PR5、NPR1、ERF1相对表达量均低于对照,而WRKY70相对表达量高于对照。GhERF5-4D能够响应枯萎病菌胁迫,并通过参与乙烯、茉莉酸途径调控下游相关抗病防卫基因提高棉花对枯萎病抗性。
赵曾强, 郭文婷, 张析, 李潇玲, 张薇. 棉花抗枯萎病相关基因GhERF5-4D的克隆及功能分析[J]. 生物技术通报, 2022, 38(4): 193-201.
ZHAO Zeng-qiang, GUO Wen-ting, ZHANG Xi, LI Xiao-ling, ZHANG Wei. Cloning and Functional Analysis of GhERF5-4D Gene Related to Fusarium oxysporum Resistance in Cotton[J]. Biotechnology Bulletin, 2022, 38(4): 193-201.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| KpnI-GhERF5-4D-F | GGGGTACCATGGCTTCTTTCGGAGA |
| BamHI-GhERF5-4D-R | CGGGATCCCATAACGGCGAGTCCTG |
| qGhERF5-4D-F | AATCCACCCTAAGCCAACGTC |
| qGhERF5-4D-R | CTTTTCTCGTCATGTTCCTCGAT |
| GhUBQ7-F | GAAGACCTACACCAAGCCCAA |
| GhUBQ7-R | CGGACTCTACTCAATCCCCACC |
| XbaI-GhERF5-4D-CO-F | GCTCTAGATCCCACCCACAAGTTTTAACAT |
| KpnI-GhERF5-4D-CO-R | CGGGTACCCTCCTTTTCCCCGACTCTG |
| GhERF1-F | CTTTAGATCACAACTCGCTTC |
| GhERF1-R | TTTTCTTTCGTCGGGCTT |
| GhPR1-F | TTTAGTAAGGTTTTTGACCGACGAA |
| GhPR1-R | GACTCTGTCCATCTTGGTTGTGCTA |
| GhPR2-F | TTCAAGGTTTGCACTCGGAAGA |
| GhPR2-R | CCACCAGCAGCAGAAGTTATCG |
| GhPR4-F | CTTGATATAAGATTGGTTAGCCC |
| GhPR4-R | TTTGCTTTTGTTGTATGTAGTCG |
| GhPR5-F | TTGGCTCTTACTTCCGACCATCT |
| GhPR5-R | GCCGTGATTCATACAGTTATCCTCA |
| GhWRKY70-F | TGGCAACATACGAAGGACAACACA |
| GhWRKY70-R | CTTCGCTCGTCCCGGAAAATATCGC |
| GhNPR1-F | CACGTGGTGCTGTTGTTGTTACTG |
| GhNPR1-R | GCGAATCGTTGCTTTCTTCTTCA |
表1 引物列表
Table 1 Table of primers
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| KpnI-GhERF5-4D-F | GGGGTACCATGGCTTCTTTCGGAGA |
| BamHI-GhERF5-4D-R | CGGGATCCCATAACGGCGAGTCCTG |
| qGhERF5-4D-F | AATCCACCCTAAGCCAACGTC |
| qGhERF5-4D-R | CTTTTCTCGTCATGTTCCTCGAT |
| GhUBQ7-F | GAAGACCTACACCAAGCCCAA |
| GhUBQ7-R | CGGACTCTACTCAATCCCCACC |
| XbaI-GhERF5-4D-CO-F | GCTCTAGATCCCACCCACAAGTTTTAACAT |
| KpnI-GhERF5-4D-CO-R | CGGGTACCCTCCTTTTCCCCGACTCTG |
| GhERF1-F | CTTTAGATCACAACTCGCTTC |
| GhERF1-R | TTTTCTTTCGTCGGGCTT |
| GhPR1-F | TTTAGTAAGGTTTTTGACCGACGAA |
| GhPR1-R | GACTCTGTCCATCTTGGTTGTGCTA |
| GhPR2-F | TTCAAGGTTTGCACTCGGAAGA |
| GhPR2-R | CCACCAGCAGCAGAAGTTATCG |
| GhPR4-F | CTTGATATAAGATTGGTTAGCCC |
| GhPR4-R | TTTGCTTTTGTTGTATGTAGTCG |
| GhPR5-F | TTGGCTCTTACTTCCGACCATCT |
| GhPR5-R | GCCGTGATTCATACAGTTATCCTCA |
| GhWRKY70-F | TGGCAACATACGAAGGACAACACA |
| GhWRKY70-R | CTTCGCTCGTCCCGGAAAATATCGC |
| GhNPR1-F | CACGTGGTGCTGTTGTTGTTACTG |
| GhNPR1-R | GCGAATCGTTGCTTTCTTCTTCA |

图1 GhERF5-4D的PCR扩增及序列分析 A:GhERF5-4D的cDNA序列及推定氨基酸序列(方框代表起始密码子ATG和终止密码子TAA,*为终止密码子,红色框为保守域结构域);B:GhERF5-4D的PCR扩增产物;C:GhERF5-4D酶切验证;D:GhERF5-4D蛋白结构域分析;E:GhERF5-4D启动子序列分析
Fig.1 PCR amplification and sequence analysis of GhERF5-4 D gene A:The cDNA sequence and the putative amino acid sequence of the GhERF5-4D(The square frame refers to initiation codon ATG and termination codon TAA,* indicates termination codon,and red square frame indicates conserved domain). B:PCR amplification result of GhERF5-4D. C:Enzyme digesting verification of GhERF5-4D. D:Analysis of structure domain of GhERF5-4D protein. E:Sequence analysis of GhERF5-4D gene promoter

图2 棉花中枯萎病菌诱导下GhERF5-4D的表达模式 A:中棉所12;B:新陆早7。不同字母表示P=0.05水平时差异显著,下同
Fig.2 Expression pattern of GhERF5-4 D gene after treat by Fusarium oxysporum in cotton A:Zhongmiansuo 12. B:Xinluzao 7. Different letters indicate significant difference at P = 0.05 level. The same below
图4 pTRV2-GhERF5-4D-CO载体的构建 A:GhERF5-4D-CO片段的扩增;B:GhERF5-4D-CO片段的酶切验证; C:pTRV2-GhERF5-4D-CO双酶切验证;D:pTRV2-GhERF5-4D-CO菌液PCR验证;M:DL2000
Fig.4 Vector construction of pTRV2-GhERF5-4D-CO A:PCR amplification of GhERF5-4D-CO. B:Enzyme digestion result of GhERF5-4D-CO. C:Enzyme digesting verification of pTRV2-GhERF5-4D-CO. D:PCR amplification result of pTRV2-GhERF5-4D-CD of agrobacterium. M:DL2000
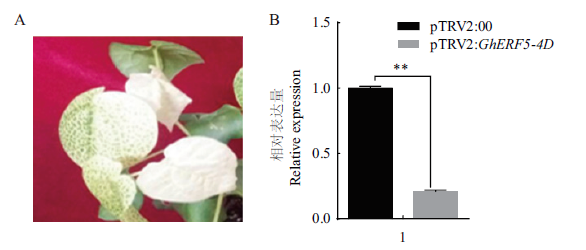
图5 GhERF5-4D沉默效率与叶片漂白情况 A:注射GhCLA1后叶片漂白情况;B:GhERF5-4D沉默效率。**表示P=0.01水平差异显著
Fig.5 GhERF5-4D gene silencing efficiency and leaf bleaching A:Leaf bleaching after the injection of GhCLA1 gene. B:GhERF5-4D gene silencing efficiency. ** indicates significant difference at P=0.01

图6 GhERF5-4D沉默后接菌处理 A:沉默植株接菌12 d后的表型;B:沉默植株接菌20 d后的表型;C:接菌20 d后棉花茎段在PDA培养基培养
Fig.6 GhERF5-4 D gene silencing followed by bacterial treatment A:The incidence phenotype of silencing plants at 12 d. B:The incidence phenot-ype of silencing plants at 20 d. C:20 d stem sections were plated on PDA medium
| [1] | 中华人民共和国统计局. 国家统计局关于2020年棉花产量的公告[EB/OL].(2020-12-18)[2021-12-06]. http://www.stats.gov.cn/tjsj/zxfb/202012/t20201218_1810113.html. |
| [2] | 李菲, 龚记熠, 李欲柯, 等. 抗旱耐盐植物功能基因发掘及其在棉花育种中的应用[J]. 分子植物育种, 2019, 17(22):7395-7400. |
| Li F, Gong JY, Li YK, et al. Excavation of drought and salt responsive functional genes and their application in cotton breeding[J]. Mol Plant Breed, 2019, 17(22):7395-7400. | |
| [3] | 李潇玲, 张析, 郭文婷, 等. 棉花抗枯萎病相关转录因子基因的克隆与表达[J]. 西北植物学报, 2017, 37(1):67-73. |
| Li XL, Zhang X, Guo WT, et al. Cloning and expression of a transcription factor gene related to cotton resistance to Fusarium oxysporum f. sp. vasinfectum[J]. Acta Bot Boreali Occidentalia Sin, 2017, 37(1):67-73. | |
| [4] |
Nakano T, Suzuki K, Fujimura T, et al. Genome-wide analysis of the ERF gene family in Arabidopsis and rice[J]. Plant Physiol, 2006, 140(2):411-432.
doi: 10.1104/pp.105.073783 URL |
| [5] |
Zhuang J, Cai B, Peng RH, et al. Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa[J]. Biochem Biophys Res Commun, 2008, 371(3):468-474.
doi: 10.1016/j.bbrc.2008.04.087 URL |
| [6] |
Licausi F, Giorgi FM, Zenoni S, et al. Genomic and transcriptomic analysis of the AP2/ERF superfamily in Vitis vinifera[J]. BMC Genomics, 2010, 11:719.
doi: 10.1186/1471-2164-11-719 URL |
| [7] |
Hu L, Liu S. Genome-wide identification and phylogenetic analysis of the ERF gene family in cucumbers[J]. Genet Mol Biol, 2011, 34(4):624-633.
doi: 10.1590/S1415-47572011005000054 URL |
| [8] |
Wessler SR. Homing into the origin of the AP2 DNA binding domain[J]. Trends Plant Sci, 2005, 10(2):54-56.
pmid: 15708341 |
| [9] |
Sakuma Y, Liu Q, Dubouzet JG, et al. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression[J]. Biochem Biophys Res Commun, 2002, 290(3):998-1009.
doi: 10.1006/bbrc.2001.6299 URL |
| [10] | 何兰兰. 棉花ERF亚族转录因子基因的克隆与特征分析[D]. 石河子:石河子大学, 2014. |
| He LL. Cloning and characterization analysis of ERF sub-family transcription factor gene from cotton(Gossypium hirsutum L.)[D]. Shihezi:Shihezi University, 2014. | |
| [11] |
Dubouzet JG, Sakuma Y, Ito Y, et al. OsDREBgenes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression[J]. Plant J, 2003, 33(4):751-763.
pmid: 12609047 |
| [12] |
Gutterson N, Reuber TL. Regulation of disease resistance pathways by AP2/ERF transcription factors[J]. Curr Opin Plant Biol, 2004, 7(4):465-471.
pmid: 15231271 |
| [13] |
McGrath KC, Dombrecht B, Manners JM, et al. Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression[J]. Plant Physiol, 2005, 139(2):949-959.
pmid: 16183832 |
| [14] |
Berrocal-Lobo M, Molina A, Solano R. Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi[J]. Plant J, 2002, 29(1):23-32.
pmid: 12060224 |
| [15] |
Pré M, Atallah M, Champion A, et al. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense[J]. Plant Physiol, 2008, 147(3):1347-1357.
doi: 10.1104/pp.108.117523 URL |
| [16] |
Oñate-Sánchez L, Anderson JP, Young J, et al. AtERF14, a member of the ERF family of transcription factors, plays a nonredundant role in plant defense[J]. Plant Physiol, 2007, 143(1):400-409.
doi: 10.1104/pp.106.086637 URL |
| [17] |
Park JM, Park CJ, Lee SB, et al. Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco[J]. Plant Cell, 2001, 13(5):1035-1046.
pmid: 11340180 |
| [18] | Fischer U, Dröge-Laser W. Overexpression of NtERF5, a new member of the tobacco ethylene response transcription factor family enhances resistance to tobacco mosaic virus[J]. Mol Plant Microbe Interactions®, 2004, 17(10):1162-1171. |
| [19] |
Chen X, Guo Z. Tobacco OPBP1 enhances salt tolerance and disease resistance of transgenic rice[J]. Int J Mol Sci, 2008, 9(12):2601-2613.
doi: 10.3390/ijms9122601 URL |
| [20] |
Gu YQ, Wildermuth MC, Chakravarthy S, et al. Tomato transcription factors pti4, pti5, and pti6 activate defense responses when expressed in Arabidopsis[J]. Plant Cell, 2002, 14(4):817-831.
doi: 10.1105/tpc.000794 URL |
| [21] |
Dong N, Liu X, Lu Y, et al. Overexpression of TaPIEP1, a pathogen-induced ERF gene of wheat, confers host-enhanced resistance to fungal pathogen Bipolaris sorokiniana[J]. Funct Integr Genomics, 2010, 10(2):215-226.
doi: 10.1007/s10142-009-0157-4 URL |
| [22] |
Jung J, Won SY, Suh SC, et al. The barley ERF-type transcription factor HvRAF confers enhanced pathogen resistance and salt tolerance in Arabidopsis[J]. Planta, 2007, 225(3):575-588.
doi: 10.1007/s00425-006-0373-2 URL |
| [23] |
Zhang G, Chen M, Li L, et al. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco[J]. J Exp Bot, 2009, 60(13):3781-3796.
doi: 10.1093/jxb/erp214 URL |
| [24] |
Cao YF, Wu YF, Zheng Z, et al. Overexpression of the rice EREBP-like gene OsBIERF3 enhances disease resistance and salt tolerance in transgenic tobacco[J]. Physiol Mol Plant Pathol, 2005, 67(3/4/5):202-211.
doi: 10.1016/j.pmpp.2006.01.004 URL |
| [25] |
Anderson JP, Lichtenzveig J, Gleason C, et al. The B-3 ethylene response factor MtERF1-1 mediates resistance to a subset of root pathogens in Medicago truncatula without adversely affecting symbiosis with rhizobia[J]. Plant Physiol, 2010, 154(2):861-873.
doi: 10.1104/pp.110.163949 URL |
| [26] |
Jin LG, Liu JY. Molecular cloning, expression profile and promoter analysis of a novel ethylene responsive transcription factor gene GhERF4 from cotton(Gossypium hirstum)[J]. Plant Physiol Biochem, 2008, 46(1):46-53.
doi: 10.1016/j.plaphy.2007.10.004 URL |
| [27] |
Jin LG, Huang B, Li H, et al. Expression profiles and transactivation analysis of a novel ethylene-responsive transcription factor gene GhERF5 from cotton[J]. Prog Nat Sci, 2009, 19(5):563-572.
doi: 10.1016/j.pnsc.2008.05.036 URL |
| [28] | Jin LG, Li H, Liu JY. Molecular characterization of three ethylene responsive element binding factor genes from cotton[J]. J Integr Plant Biol, 2010, 52(5):485-495. |
| [29] |
Zuo KJ, Qin J, Zhao JY, et al. Over-expression GbERF2 transcription factor in tobacco enhances brown spots disease resistance by activating expression of downstream genes[J]. Gene, 2007, 391(1/2):80-90.
doi: 10.1016/j.gene.2006.12.019 URL |
| [30] | 韩泽刚, 赵曾强, 李会会, 等. 枯萎病菌诱导感、抗陆地棉品种的基因表达谱分析[J]. 作物学报, 2015, 41(8):1201-1211. |
|
Han ZG, Zhao ZQ, Li HH, et al. Expression profiling analysis between resistant and susceptible cotton cultivars(Gossypium hirsutum L.)in response to Fusarium wilt[J]. Acta Agron Sin, 2015, 41(8):1201-1211.
doi: 10.3724/SP.J.1006.2015.01201 URL |
|
| [31] | 韩泽刚, 赵曾强, 何兰兰, 等. 枯萎病菌诱导感、抗陆地棉品种的转录因子表达变化[J]. 作物学报, 2015, 41(2):228-239. |
|
Han ZG, Zhao ZQ, He LL, et al. Expression changes of transcription factors in susceptible and resistant upland cotton(Gossypium hirsutum L.)cultivars in response to Fusarium wilt[J]. Acta Agron Sin, 2015, 41(2):228-239.
doi: 10.3724/SP.J.1006.2015.00228 URL |
|
| [32] |
Finatto T, Viana VE, Woyann LG, et al. Can WRKY transcription factors help plants to overcome environmental challenges?[J]. Genet Mol Biol, 2018, 41(3):533-544.
doi: 10.1590/1678-4685-gmb-2017-0232 URL |
| [33] |
Baillo, Kimotho, Zhang, et al. Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement[J]. Genes, 2019, 10(10):771.
doi: 10.3390/genes10100771 URL |
| [34] |
Xie Z, Nolan TM, Jiang H, et al. AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis[J]. Front Plant Sci, 2019, 10:228.
doi: 10.3389/fpls.2019.00228 URL |
| [35] |
Li P, Chai Z, Lin P, et al. Genome-wide identification and expression analysis of AP2/ERF transcription factors in sugarcane(Saccharum spontaneum L.)[J]. BMC Genomics, 2020, 21(1):685.
doi: 10.1186/s12864-020-07076-x URL |
| [36] |
Guo W, Jin L, Miao Y, et al. An ethylene response-related factor, GbERF1-like, from Gossypium barbadense improves resistance to Verticillium dahliae via activating lignin synjournal[J]. Plant Mol Biol, 2016, 91(3):305-318.
doi: 10.1007/s11103-016-0467-6 URL |
| [37] |
Cacas JL, Pré M, Pizot M, et al. GhERF-IIb3 regulates the accumulation of jasmonate and leads to enhanced cotton resistance to blight disease[J]. Mol Plant Pathol, 2017, 18(6):825-836.
doi: 10.1111/mpp.12445 URL |
| [38] |
Caarls L, Van der Does D, Hickman R, et al. Assessing the role of ETHYLENE RESPONSE FACTOR transcriptional repressors in salicylic acid-mediated suppression of jasmonic acid-responsive genes[J]. Plant Cell Physiol, 2017, 58(2):266-278.
doi: 10.1093/pcp/pcw187 pmid: 27837094 |
| [39] |
Champion A, Hebrard E, Parra B, et al. Molecular diversity and gene expression of cotton ERF transcription factors reveal that group IXa members are responsive to jasmonate, ethylene and And[J]. Mol Plant Pathol, 2009, 10(4):471-485.
doi: 10.1111/j.1364-3703.2009.00549.x pmid: 19523101 |
| [40] |
Liu J, Wang Y, Zhao G, et al. A novel Gossypium barbadense ERF transcription factor, GbERFb, regulation host response and resistance to Verticillium dahliae in tobacco[J]. Physiol Mol Biol Plants, 2017, 23(1):125-134.
doi: 10.1007/s12298-016-0402-y URL |
| [41] |
Dong L, Cheng Y, Wu J, et al. Overexpression of GmERF5, a new member of the soybean EAR motif-containing ERF transcription factor, enhances resistance to Phytophthora sojae in soybean[J]. J Exp Bot, 2015, 66(9):2635-2647.
doi: 10.1093/jxb/erv078 URL |
| [42] | 金莉. 棉花GbERF1基因的克隆与功能鉴定[D]. 武汉:华中农业大学, 2012. |
| Jin L. Isolation and characterization of GbERF1 in cotton[D]. Wuhan:Huazhong Agricultural University, 2012. | |
| [43] | Moffat CS, Ingle RA, Wathugala DL, et al. ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinerea in Arabidopsis[J]. PLoS One, 2012, 7(4):e35995. |
| [44] | 李文正, 张海文, 王俊英, 等. ERF转录因子及其在烟草抗逆性改良中的应用[J]. 生物技术通报, 2006(4):30-34. |
| Li WZ, Zhang HW, Wang JY, et al. Ethylene responsive factors and their application in improving tobacco tolerance to biotic and abiotic stresses[J]. Biotechnol Bull, 2006(4):30-34. |
| [1] | 娄慧, 朱金成, 杨洋, 张薇. 抗、感品种棉花根系分泌物对尖孢镰刀菌生长及基因表达的影响[J]. 生物技术通报, 2023, 39(9): 156-167. |
| [2] | 王佳蕊, 孙培媛, 柯瑾, 冉彬, 李洪有. 苦荞糖基转移酶基因FtUGT143的克隆及表达分析[J]. 生物技术通报, 2023, 39(8): 204-212. |
| [3] | 孙明慧, 吴琼, 刘丹丹, 焦小雨, 王文杰. 茶树CsTMFs的克隆与表达分析[J]. 生物技术通报, 2023, 39(7): 151-159. |
| [4] | 赵雪婷, 高利燕, 王俊刚, 沈庆庆, 张树珍, 李富生. 甘蔗AP2/ERF转录因子基因ShERF3的克隆、表达及其编码蛋白的定位[J]. 生物技术通报, 2023, 39(6): 208-216. |
| [5] | 杨洋, 朱金成, 娄慧, 韩泽刚, 张薇. 海岛棉与枯萎病菌的互作转录组分析[J]. 生物技术通报, 2023, 39(6): 259-273. |
| [6] | 姜晴春, 杜洁, 王嘉诚, 余知和, 王允, 柳忠玉. 虎杖转录因子PcMYB2的表达特性和功能分析[J]. 生物技术通报, 2023, 39(5): 217-223. |
| [7] | 姚姿婷, 曹雪颖, 肖雪, 李瑞芳, 韦小妹, 邹承武, 朱桂宁. 火龙果溃疡病菌实时荧光定量PCR内参基因的筛选[J]. 生物技术通报, 2023, 39(5): 92-102. |
| [8] | 王艺清, 王涛, 韦朝领, 戴浩民, 曹士先, 孙威江, 曾雯. 茶树SMAS基因家族的鉴定及互作分析[J]. 生物技术通报, 2023, 39(4): 246-258. |
| [9] | 刘思佳, 王浩楠, 付宇辰, 闫文欣, 胡增辉, 冷平生. ‘西伯利亚’百合LiCMK基因克隆及功能分析[J]. 生物技术通报, 2023, 39(3): 196-205. |
| [10] | 王涛, 漆思雨, 韦朝领, 王艺清, 戴浩民, 周喆, 曹士先, 曾雯, 孙威江. CsPPR和CsCPN60-like在茶树白化叶片中的表达分析及互作蛋白验证[J]. 生物技术通报, 2023, 39(3): 218-231. |
| [11] | 庞强强, 孙晓东, 周曼, 蔡兴来, 张文, 王亚强. 菜心BrHsfA3基因克隆及其对高温胁迫的响应[J]. 生物技术通报, 2023, 39(2): 107-115. |
| [12] | 苗淑楠, 高宇, 李昕儒, 蔡桂萍, 张飞, 薛金爱, 季春丽, 李润植. 大豆GmPDAT1参与油脂合成和非生物胁迫应答的功能分析[J]. 生物技术通报, 2023, 39(2): 96-106. |
| [13] | 葛雯冬, 王腾辉, 马天意, 范震宇, 王玉书. 结球甘蓝PRX基因家族全基因组鉴定与逆境条件下的表达分析[J]. 生物技术通报, 2023, 39(11): 252-260. |
| [14] | 杨旭妍, 赵爽, 马天意, 白玉, 王玉书. 三个甘蓝WRKY基因的克隆及其对非生物胁迫的表达[J]. 生物技术通报, 2023, 39(11): 261-269. |
| [15] | 陈楚怡, 杨小梅, 陈胜艳, 陈斌, 岳莉然. ABA和干旱胁迫下菊花脑ZF-HD基因家族的表达分析[J]. 生物技术通报, 2023, 39(11): 270-282. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||