








生物技术通报 ›› 2022, Vol. 38 ›› Issue (4): 202-216.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0830
镐青青( ), 姚圣, 刘佳禾, 陈佩珍, 张梦洋, 季孔庶(
), 姚圣, 刘佳禾, 陈佩珍, 张梦洋, 季孔庶( )
)
收稿日期:2021-06-30
出版日期:2022-04-26
发布日期:2022-05-06
通讯作者:
季孔庶,男,博士,教授,研究方向:林木遗传育种、园林植物遗传育种;E-mail: ksji@njfu.edu.cn作者简介:镐青青,女,硕士研究生,研究方向:林木遗传育种;E-mail: hqq@njfu.edu.cn
基金资助:
HAO Qing-qing( ), YAO Sheng, LIU Jia-he, CHEN Pei-zhen, ZHANG Meng-yang, JI Kong-shu(
), YAO Sheng, LIU Jia-he, CHEN Pei-zhen, ZHANG Meng-yang, JI Kong-shu( )
)
Received:2021-06-30
Published:2022-04-26
Online:2022-05-06
摘要:
NAC(NAM,ATAF1/2,CUC2)家族基因参与调控植物生长发育、响应非生物胁迫及激素信号转导。研究马尾松中NAC转录因子在非生物胁迫中的应答,为阐明基因功能提供理论依据。以马尾松(Pinus massoniana)为试验材料,通过RT-PCR及RACE技术克隆一个NAC家族基因,并对其进行生物信息学分析。运用qRT-PCR检测其组织表达特性及对逆境胁迫的响应情况;以基因组DNA为模板,运用染色体步移(genome walking)技术克隆该基因上游启动子序列;通过农杆菌注射本氏烟草(Nicotiana benthamiana),观察其亚细胞定位。该基因全长1 726 bp,开放阅读框为1 209 bp,编码402个氨基酸,将其命名为PmNAC8,GenBank登录号为MZ291447;亚细胞定位结果显示,该基因定位于细胞核;其密码子使用偏性较弱,偏好使用A/T结尾的密码子,且烟草与酵母更适合作为PmNAC8的异源表达受体;qRT-PCR显示PmNAC8在根部表达量最高,且受机械损伤及植物激素GA、ABA的诱导;克隆获得PmNAC8上游1 492 bp的启动子区域,含有多个胁迫诱导元件、赤霉素调控元件和光响应元件。PmNAC8参与多种逆境胁迫,可能在赤霉素信号途径中发挥重要作用。
镐青青, 姚圣, 刘佳禾, 陈佩珍, 张梦洋, 季孔庶. 马尾松NAC转录因子基因PmNAC8的克隆及表达分析[J]. 生物技术通报, 2022, 38(4): 202-216.
HAO Qing-qing, YAO Sheng, LIU Jia-he, CHEN Pei-zhen, ZHANG Meng-yang, JI Kong-shu. Cloning and Expression Analysis of NAC Transcription Factor PmNAC8 in Pinus massoniana[J]. Biotechnology Bulletin, 2022, 38(4): 202-216.
| 引物名称 Primer name | 引物序列 Primer sequence (5'-3') | 用途 Purpose |
|---|---|---|
| PmNAC8-Mid F | GAAGGTATGTGAAAATGGGG | 中间片段扩增前引物 Forward primer of middle fragment amplification |
| PmNAC8-Mid R | GGGAATCTCAGTGTTTTGAAG | 中间片段扩增后引物 Reverse primer of middle fragment amplification reverse primer |
| 3' RACE-PmNAC8 F | GGCAGGGGAATCTCAGTGTTTTGAAG | 3'-巢氏PCR前引物 Forward primer of 3'-nested PCR |
| 3' RACE-PmNAC8 R | TACCGTCGTTCCACTAGTGATTT | 3'-巢氏PCR后引物 Reverse primer of 3'-nested PCR |
| 5' RACE-PmNAC8 F | CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT | 5'-巢氏PCR前引物 Forward primer of 5'-nested PCR |
| 5' RACE-PmNAC8 R | CAATACCATCTTGGGACTGAGGACGA | 5'-巢氏PCR后引物 Reverse primer of 5'-nested PCR |
| PmNAC8-F | CAAAGTGGAAACTCTGAGCAGAGC | 全长扩增前引物 Forward primer of total length of amplification |
| PmNAC8-R | TTCCGAAGACGGCAAAAATAGTTG | 全长扩增后引物 Reverse primer of total length of amplification |
| PmNAC8-ORF F | ATGGCGAGACCATGGATTACTG | 编码阅读框前引物 Forward primer of code reading frame forward primer |
| PmNAC8-ORF R | TTGGCTGTTACCAAAGTTCTTTAACC | 编码阅读框后引物 Reverse primer of code reading frame forward primer |
| Tubulin alpha F | CACAGAAAGCTGCTCATGGTAA | 内参基因前引物 Forward primer of internal genes |
| Tubulin alpha R | CTGCGACTATGAGTGGAGTGA | 内参基因后引物 Reverse primer of internal genes |
| Q-PmNAC8 F | TCTGATTGTGGTCGGATTGGAGG | 定量及半定量前引物 Forward primer of the quantitative and semi-quantitative |
| Q-PmNAC8 R | AGGCTTTCTTGGGAACTGAACTG | 定量及半定量后引物 Reverse primer of the quantitative and semi-quantitative |
| PmNAC8-GFP F | gagaacacgggggactctagaATGGCGAGACCATGGATTACTG | 重组表达载体前引物 Forward primer of recombinant expression vector |
| PmNAC8-GFP R | gcccttgctcaccatggatccTTGGCTGTTACCAAAGTTCTTTAACC | 重组表达载体后引物 Reverse primer of recombinant expression vector |
| GSP1 | TTGCGATTCTCCTACCATCAGTAA | 启动子扩增特异性引物1 The promoter amplification specificity primer 1 |
| GSP2 | ACATCATTTCTCCCGTTCTCGTTT | 启动子扩增特异性引物2 The promoter amplification specificity primer 2 |
| PmNAC8-Pro F | TTTGGTTAGCGTAGAGGGTTGTCA | 启动子全长克隆前引物 Forward primer of total length of the promoter cloning |
| PmNAC8-Pro R | CCGTGTCGCCTTACATCATTTCTC | 启动子全长克隆后引物 Reverse primer of total length of the promoter cloning |
表1 本研究中使用的引物序列
Table 1 Sequences of primers used in this study
| 引物名称 Primer name | 引物序列 Primer sequence (5'-3') | 用途 Purpose |
|---|---|---|
| PmNAC8-Mid F | GAAGGTATGTGAAAATGGGG | 中间片段扩增前引物 Forward primer of middle fragment amplification |
| PmNAC8-Mid R | GGGAATCTCAGTGTTTTGAAG | 中间片段扩增后引物 Reverse primer of middle fragment amplification reverse primer |
| 3' RACE-PmNAC8 F | GGCAGGGGAATCTCAGTGTTTTGAAG | 3'-巢氏PCR前引物 Forward primer of 3'-nested PCR |
| 3' RACE-PmNAC8 R | TACCGTCGTTCCACTAGTGATTT | 3'-巢氏PCR后引物 Reverse primer of 3'-nested PCR |
| 5' RACE-PmNAC8 F | CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT | 5'-巢氏PCR前引物 Forward primer of 5'-nested PCR |
| 5' RACE-PmNAC8 R | CAATACCATCTTGGGACTGAGGACGA | 5'-巢氏PCR后引物 Reverse primer of 5'-nested PCR |
| PmNAC8-F | CAAAGTGGAAACTCTGAGCAGAGC | 全长扩增前引物 Forward primer of total length of amplification |
| PmNAC8-R | TTCCGAAGACGGCAAAAATAGTTG | 全长扩增后引物 Reverse primer of total length of amplification |
| PmNAC8-ORF F | ATGGCGAGACCATGGATTACTG | 编码阅读框前引物 Forward primer of code reading frame forward primer |
| PmNAC8-ORF R | TTGGCTGTTACCAAAGTTCTTTAACC | 编码阅读框后引物 Reverse primer of code reading frame forward primer |
| Tubulin alpha F | CACAGAAAGCTGCTCATGGTAA | 内参基因前引物 Forward primer of internal genes |
| Tubulin alpha R | CTGCGACTATGAGTGGAGTGA | 内参基因后引物 Reverse primer of internal genes |
| Q-PmNAC8 F | TCTGATTGTGGTCGGATTGGAGG | 定量及半定量前引物 Forward primer of the quantitative and semi-quantitative |
| Q-PmNAC8 R | AGGCTTTCTTGGGAACTGAACTG | 定量及半定量后引物 Reverse primer of the quantitative and semi-quantitative |
| PmNAC8-GFP F | gagaacacgggggactctagaATGGCGAGACCATGGATTACTG | 重组表达载体前引物 Forward primer of recombinant expression vector |
| PmNAC8-GFP R | gcccttgctcaccatggatccTTGGCTGTTACCAAAGTTCTTTAACC | 重组表达载体后引物 Reverse primer of recombinant expression vector |
| GSP1 | TTGCGATTCTCCTACCATCAGTAA | 启动子扩增特异性引物1 The promoter amplification specificity primer 1 |
| GSP2 | ACATCATTTCTCCCGTTCTCGTTT | 启动子扩增特异性引物2 The promoter amplification specificity primer 2 |
| PmNAC8-Pro F | TTTGGTTAGCGTAGAGGGTTGTCA | 启动子全长克隆前引物 Forward primer of total length of the promoter cloning |
| PmNAC8-Pro R | CCGTGTCGCCTTACATCATTTCTC | 启动子全长克隆后引物 Reverse primer of total length of the promoter cloning |

图1 马尾松PmNAC8的全长及ORF的扩增 A:马尾松PmNAC8基因cDNA全长凝胶电泳图;B:马尾松PmNAC8基因ORF凝胶电泳图;M:DL2502 DNA marker
Fig. 1 Full-length and ORF amplification of PmNAC8 gene in P. massoniana A:Electropherogram of full-length PmNAC8 cDNA of P. massoniana. B:Electropherogram of ORF of PmNAC8 of P. massoniana. M:DL2502 DNA marker
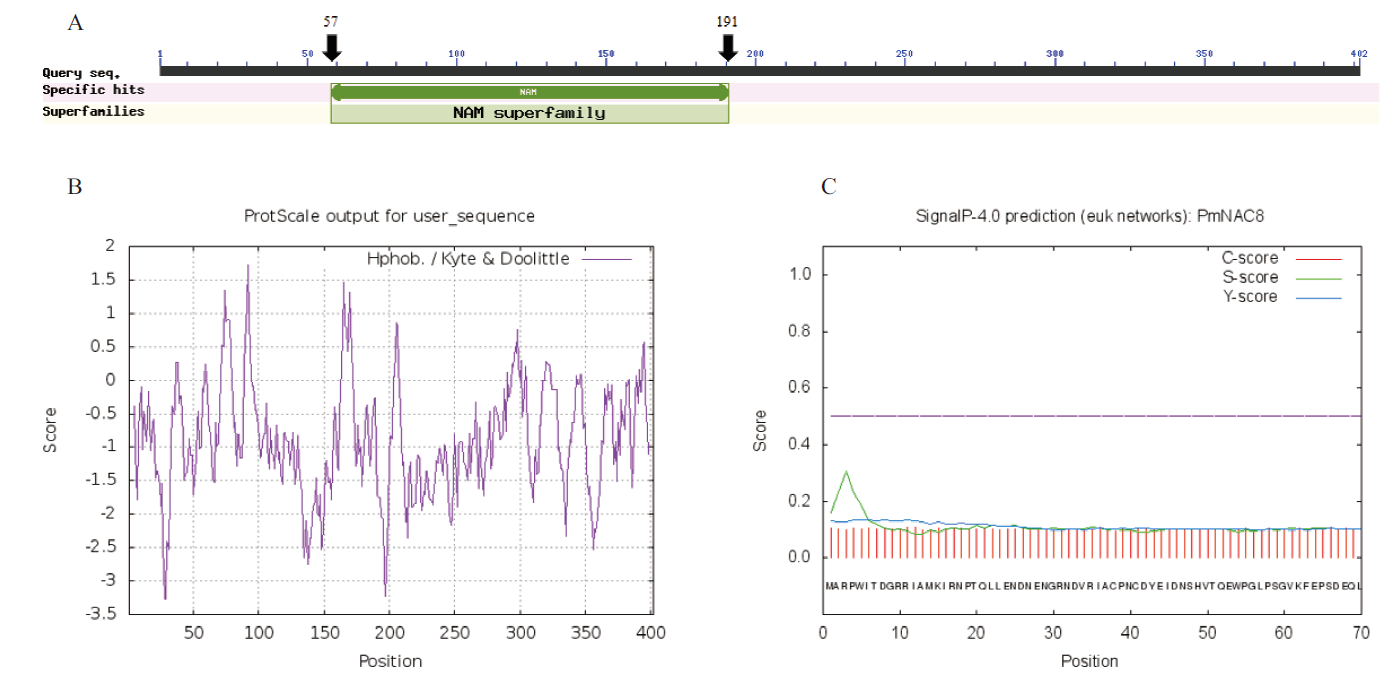
图2 PmNAC8蛋白的生物信息学分析 A:PmNAC8蛋白的保守结构域分析;B:PmNAC8蛋白疏水性的预测分析;C:PmNAC8蛋白的信号肽预测分析
Fig.2 Bioinformatics analysis of PmNAC8 protein A:Conserved domain prediction of PmNAC8 protein. B:Hydrophobicity prediction of PmNAC8 protein. C:Signal peptide prediction of PmNAC8 protein

图5 PmNAC8蛋白系统发育进化树 火炬松Pinus taeda,AZA14807.1;北美云杉Picea sitchensis,ABR16510.1;互叶梅Amborella trichopoda,XP_011627944.1;苎麻Boehmeria nivea,AIA57535.1;茶树Camellia sinensis,XP_028087922.1,XP_028114758.1;薄壳山核桃Carya illinoinensis,KAG2710462.1;扁嘴豆Cicer arietinum,XP_004490275.1,XP_004490275.1;香瓜Cucumis melo,XP_008446555.1;大豆Glycine max,XP_006605344.1;陆地棉Gossypium hirsutum,NP_001314441.1;橡胶树Hevea brasiliensis,XP_021658485.1;麻风树Jatropha curcas,XP_012072809.1;博落回Macleaya cordata,OVA20812.1;木薯Manihot esculenta,ALC79049.1,XP_021594205.1;白花草木樨Melilotus albus,QSD99866.1;苦瓜Momordica charantia,XP_022150639.1;川桑Morus notabilis,XP_024019402.1,XP_024019400.1;蓝果树Nyssa sinensis,KAA8529210.1;可可Theobroma cacao,EOY02531.1,XP_007031605.2;葡萄Vitis vinifera,XP_002281522.1,XP_010647968.1,XP_002272446.1;阿月浑子Pistacia vera,XP_031266521.1;雷公藤Tripterygium wilfordii,XP_038679434.1;睡莲Nymphaea colorata,XP_031480907.1;红花烟草Nicotiana tabacum,XP_016434052.1;烟草Nicotiana attenuata,XP_019256868.1;杨梅Morella rubra,KAB1221131.1;银白杨Populus alba,XP_034902708.1;枣Ziziphus jujuba,XP_024933773.1
Fig.5 Phylogenetic analysis of PmNAC8 protein
| 物种 Species | 登录号 GenBank accession number | A3s | C3s | G3s | T3s | ENc | CAI | GC3s/% | GC/% |
|---|---|---|---|---|---|---|---|---|---|
| 马尾松 P. massoniana | MZ291447 | 0.443 | 0.182 | 0.241 | 0.451 | 50.19 | 0.187 | 31.10 | 42.50 |
表2 PmNAC8密码子选择偏好性参数
Table 2 Codon selection preference parameters of PmNAC8 gene
| 物种 Species | 登录号 GenBank accession number | A3s | C3s | G3s | T3s | ENc | CAI | GC3s/% | GC/% |
|---|---|---|---|---|---|---|---|---|---|
| 马尾松 P. massoniana | MZ291447 | 0.443 | 0.182 | 0.241 | 0.451 | 50.19 | 0.187 | 31.10 | 42.50 |
| 氨基酸 Amino acid | 密码子 Codon | 相对密码子 使用度RSCU | 个数 Number | 频率 Frequency | 比例 Fraction | 氨基酸 Amino acid | 密码子 Codon | 相对密码子 使用度RSCU | 个数 Number | 频率 Frequency | 比例 Fraction | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phe | UUU | 1.54 | 10 | 24.814 | 0.769 | Tyr | UAU | 1.71 | 6 | 14.888 | 0.857 | |
| UUC | 0.46 | 3 | 7.444 | 0.231 | UAC | 0.29 | 1 | 2.481 | 0.143 | |||
| Leu | UUA | 1.50 | 6 | 14.888 | 0.250 | TER | UAA | 0.00 | 0 | 0.000 | 0.000 | |
| UUG | 1.25 | 5 | 12.407 | 0.208 | UAG | 0.00 | 0 | 0.000 | 0.000 | |||
| CUU | 1.75 | 7 | 17.370 | 0.92 | His | CAU | 1.47 | 11 | 27.295 | 0.733 | ||
| CUC | 1.00 | 4 | 9.926 | 0.167 | CAC | 0.53 | 4 | 9.926 | 0.267 | |||
| CUA | 0.50 | 2 | 4.963 | 0.083 | Gln | CAA | 1.10 | 11 | 27.295 | 0.550 | ||
| CUG | 0.00 | 0 | 0.000 | 0.000 | CAG | 0.90 | 9 | 22.333 | 0.450 | |||
| Ile | AUU | 1.43 | 10 | 24.814 | 0.476 | Asn | AAU | 1.09 | 12 | 29.777 | 0.545 | |
| AUC | 0.57 | 4 | 9.926 | 0.191 | AAC | 0.91 | 10 | 24.814 | 0.455 | |||
| AUA | 1.00 | 7 | 17.370 | 0.333 | Lys | AAA | 1.04 | 11 | 27.295 | 0.478 | ||
| Met | AUG | 1.00 | 7 | 17.370 | 1.000 | AAG | 0.96 | 12 | 29.777 | 0.522 | ||
| Val | GUU | 1.25 | 5 | 12.407 | 0.313 | Asp | GAU | 1.57 | 22 | 54.591 | 0.786 | |
| GUC | 0.75 | 3 | 7.444 | 0.187 | GAC | 0.43 | 6 | 14.888 | 0.214 | |||
| GUA | 0.75 | 3 | 7.444 | 0.187 | Glu | GAA | 1.22 | 22 | 54.591 | 0.611 | ||
| GUG | 1.25 | 5 | 12.407 | 0.313 | GAG | 0.78 | 14 | 34.739 | 0.389 | |||
| Ser | UCU | 1.31 | 7 | 17.370 | 0.318 | Cys | UGU | 1.54 | 10 | 24.814 | 0.769 | |
| UCC | 0.38 | 2 | 4.963 | 0.091 | UGC | 0.46 | 3 | 7.444 | 0.231 | |||
| UCA | 2.44 | 13 | 32.258 | 0.591 | TER | UGA | 3.00 | 1 | 2.481 | 1.000 | ||
| UCG | 0.00 | 0 | 0.000 | 0.000 | Trp | UGG | 1.00 | 6 | 14.888 | 1.000 | ||
| Pro | CCU | 1.57 | 11 | 27.295 | 0.393 | Arg | CGU | 1.67 | 5 | 12.407 | 0.625 | |
| CCC | 0.57 | 4 | 9.926 | 0.143 | CGC | 0.00 | 0 | 0.000 | 0.000 | |||
| CCA | 1.86 | 13 | 32.258 | 0.464 | CGA | 0.67 | 2 | 4.963 | 0.250 | |||
| CCG | 0.00 | 0 | 0.000 | 0.00 | CGG | 0.33 | 1 | 2.481 | 0.125 | |||
| Thr | ACU | 1.60 | 10 | 24.814 | 0.400 | Ser | AGU | 0.56 | 3 | 7.444 | 0.300 | |
| ACC | 0.16 | 1 | 2.481 | 0.040 | AGC | 1.31 | 7 | 13.370 | 0.700 | |||
| ACA | 1.60 | 10 | 24.814 | 0.400 | Arg | AGA | 1.67 | 5 | 12.407 | 0.500 | ||
| ACG | 0.64 | 4 | 9.926 | 0.160 | AGG | 1.67 | 5 | 12.407 | 0.500 | |||
| Ala | GCU | 0.86 | 3 | 7.444 | 0.214 | Gly | GGU | 0.88 | 7 | 13.370 | 0.700 | |
| GCC | 0.29 | 1 | 2.481 | 0.071 | GGC | 0.38 | 3 | 7.444 | 0.300 | |||
| GCA | 2.57 | 9 | 22.333 | 0.643 | GGA | 1.63 | 13 | 32.258 | 0.591 | |||
| GCG | 0.29 | 1 | 2.481 | 0.072 | GGG | 1.13 | 9 | 22.333 | 0.409 |
表3 PmNAC8同义密码子相对使用度
Table 3 RSCU of PmNAC8 gene
| 氨基酸 Amino acid | 密码子 Codon | 相对密码子 使用度RSCU | 个数 Number | 频率 Frequency | 比例 Fraction | 氨基酸 Amino acid | 密码子 Codon | 相对密码子 使用度RSCU | 个数 Number | 频率 Frequency | 比例 Fraction | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phe | UUU | 1.54 | 10 | 24.814 | 0.769 | Tyr | UAU | 1.71 | 6 | 14.888 | 0.857 | |
| UUC | 0.46 | 3 | 7.444 | 0.231 | UAC | 0.29 | 1 | 2.481 | 0.143 | |||
| Leu | UUA | 1.50 | 6 | 14.888 | 0.250 | TER | UAA | 0.00 | 0 | 0.000 | 0.000 | |
| UUG | 1.25 | 5 | 12.407 | 0.208 | UAG | 0.00 | 0 | 0.000 | 0.000 | |||
| CUU | 1.75 | 7 | 17.370 | 0.92 | His | CAU | 1.47 | 11 | 27.295 | 0.733 | ||
| CUC | 1.00 | 4 | 9.926 | 0.167 | CAC | 0.53 | 4 | 9.926 | 0.267 | |||
| CUA | 0.50 | 2 | 4.963 | 0.083 | Gln | CAA | 1.10 | 11 | 27.295 | 0.550 | ||
| CUG | 0.00 | 0 | 0.000 | 0.000 | CAG | 0.90 | 9 | 22.333 | 0.450 | |||
| Ile | AUU | 1.43 | 10 | 24.814 | 0.476 | Asn | AAU | 1.09 | 12 | 29.777 | 0.545 | |
| AUC | 0.57 | 4 | 9.926 | 0.191 | AAC | 0.91 | 10 | 24.814 | 0.455 | |||
| AUA | 1.00 | 7 | 17.370 | 0.333 | Lys | AAA | 1.04 | 11 | 27.295 | 0.478 | ||
| Met | AUG | 1.00 | 7 | 17.370 | 1.000 | AAG | 0.96 | 12 | 29.777 | 0.522 | ||
| Val | GUU | 1.25 | 5 | 12.407 | 0.313 | Asp | GAU | 1.57 | 22 | 54.591 | 0.786 | |
| GUC | 0.75 | 3 | 7.444 | 0.187 | GAC | 0.43 | 6 | 14.888 | 0.214 | |||
| GUA | 0.75 | 3 | 7.444 | 0.187 | Glu | GAA | 1.22 | 22 | 54.591 | 0.611 | ||
| GUG | 1.25 | 5 | 12.407 | 0.313 | GAG | 0.78 | 14 | 34.739 | 0.389 | |||
| Ser | UCU | 1.31 | 7 | 17.370 | 0.318 | Cys | UGU | 1.54 | 10 | 24.814 | 0.769 | |
| UCC | 0.38 | 2 | 4.963 | 0.091 | UGC | 0.46 | 3 | 7.444 | 0.231 | |||
| UCA | 2.44 | 13 | 32.258 | 0.591 | TER | UGA | 3.00 | 1 | 2.481 | 1.000 | ||
| UCG | 0.00 | 0 | 0.000 | 0.000 | Trp | UGG | 1.00 | 6 | 14.888 | 1.000 | ||
| Pro | CCU | 1.57 | 11 | 27.295 | 0.393 | Arg | CGU | 1.67 | 5 | 12.407 | 0.625 | |
| CCC | 0.57 | 4 | 9.926 | 0.143 | CGC | 0.00 | 0 | 0.000 | 0.000 | |||
| CCA | 1.86 | 13 | 32.258 | 0.464 | CGA | 0.67 | 2 | 4.963 | 0.250 | |||
| CCG | 0.00 | 0 | 0.000 | 0.00 | CGG | 0.33 | 1 | 2.481 | 0.125 | |||
| Thr | ACU | 1.60 | 10 | 24.814 | 0.400 | Ser | AGU | 0.56 | 3 | 7.444 | 0.300 | |
| ACC | 0.16 | 1 | 2.481 | 0.040 | AGC | 1.31 | 7 | 13.370 | 0.700 | |||
| ACA | 1.60 | 10 | 24.814 | 0.400 | Arg | AGA | 1.67 | 5 | 12.407 | 0.500 | ||
| ACG | 0.64 | 4 | 9.926 | 0.160 | AGG | 1.67 | 5 | 12.407 | 0.500 | |||
| Ala | GCU | 0.86 | 3 | 7.444 | 0.214 | Gly | GGU | 0.88 | 7 | 13.370 | 0.700 | |
| GCC | 0.29 | 1 | 2.481 | 0.071 | GGC | 0.38 | 3 | 7.444 | 0.300 | |||
| GCA | 2.57 | 9 | 22.333 | 0.643 | GGA | 1.63 | 13 | 32.258 | 0.591 | |||
| GCG | 0.29 | 1 | 2.481 | 0.072 | GGG | 1.13 | 9 | 22.333 | 0.409 |
| 氨基酸 Amino acid | 密码子 Codon | 马尾松PmNAC8 | 拟南芥基因组At | 烟草基因组Nt | 酵母基因组Sc | 大肠杆菌基因组Ec | 欧洲山杨基因组Pt | PmNAC8/At | PmNAC8/Nt | PmNAC8/Sc | PmNAC8/Ec | PmNAC8/Pt |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phe | UUU | 24.814 | 21.8 | 25.1 | 26.1 | 22.2 | 28.2 | 1.14 | 0.99 | 0.95 | 1.12 | 0.10 |
| UUC | 7.444 | 20.7 | 18.0 | 18.4 | 15.9 | 17.3 | 0.36 | 0.36 | 0.40 | 0.47 | 0.43 | |
| Leu | UUA | 14.888 | 12.7 | 13.4 | 26.2 | 13.8 | 14.0 | 1.17 | 1.11 | 0.57 | 1.08 | 1.06 |
| UUG | 12.407 | 20.9 | 22.3 | 27.2 | 13.0 | 26.3 | 0.59 | 0.56 | 0.55 | 0.95 | 0.47 | |
| CUU | 17.370 | 24.1 | 24.0 | 12.3 | 11.4 | 22.7 | 0.72 | 0.72 | 1.41 | 1.52 | 0.77 | |
| CUC | 9.926 | 16.1 | 12.3 | 5.4 | 10.5 | 15.6 | 0.62 | 0.81 | 1.84 | 0.95 | 0.64 | |
| CUA | 4.963 | 9.9 | 9.4 | 13.4 | 3.9 | 7.6 | 0.50 | 0.53 | 0.37 | 1.27 | 0.65 | |
| CUG | 0.000 | 9.8 | 10.2 | 10.5 | 51.1 | 8.5 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Ile | AUU | 24.814 | 21.5 | 27.8 | 30.1 | 29.7 | 28.1 | 1.15 | 0.89 | 0.82 | 0.84 | 0.88 |
| AUC | 9.926 | 18.5 | 13.9 | 17.2 | 23.9 | 14.5 | 0.54 | 0.71 | 0.58 | 0.42 | 0.68 | |
| AUA | 17.370 | 12.6 | 14.0 | 17.8 | 5.5 | 15.0 | 1.38 | 1.24 | 0.98 | 3.16 | 1.16 | |
| Met | AUG | 17.370 | 24.5 | 25.0 | 20.9 | 27.2 | 23.6 | 0.71 | 0.69 | 0.83 | 0.64 | 0.74 |
| Val | GUU | 12.407 | 27.2 | 26.8 | 22.1 | 18.1 | 39.1 | 0.46 | 0.46 | 0.56 | 0.69 | 0.32 |
| GUC | 7.444 | 12.8 | 11.1 | 11.8 | 14.8 | 10.8 | 0.58 | 0.67 | 0.63 | 0.50 | 0.69 | |
| GUA | 7.444 | 9.9 | 11.4 | 11.8 | 10.9 | 9.4 | 0.75 | 0.65 | 0.63 | 0.68 | 0.79 | |
| GUG | 12.407 | 17.4 | 16.7 | 10.8 | 26.2 | 19.8 | 0.72 | 0.74 | 1.15 | 0.47 | 0.63 | |
| Ser | UCU | 17.370 | 25.2 | 20.0 | 23.5 | 8.7 | 18.3 | 0.69 | 0.87 | 0.74 | 2.00 | 0.95 |
| UCC | 4.963 | 11.2 | 10.2 | 14.2 | 8.9 | 11.8 | 0.44 | 0.49 | 0.35 | 0.56 | 0.42 | |
| UCA | 32.258 | 18.3 | 17.6 | 18.7 | 8.1 | 21.2 | 1.76 | 1.83 | 1.73 | 3.98 | 1.52 | |
| UCG | 0.000 | 9.3 | 5.3 | 8.6 | 8.8 | 8.3 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Pro | CCU | 27.295 | 18.7 | 18.7 | 13.5 | 7.2 | 15.8 | 1.46 | 1.46 | 2.02 | 3.80 | 1.73 |
| CCC | 9.926 | 5.3 | 6.6 | 6.8 | 5.6 | 8.0 | 1.87 | 1.50 | 1.46 | 1.77 | 1.24 | |
| CCA | 32.258 | 16.1 | 19.8 | 18.3 | 8.4 | 16.1 | 2.00 | 1.63 | 1.76 | 3.84 | 2.00 | |
| CCG | 0.000 | 8.6 | 5.0 | 5.3 | 22.4 | 7.4 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Thr | ACU | 24.814 | 17.5 | 20.3 | 20.3 | 9.1 | 19.7 | 1.42 | 1.22 | 1.22 | 2.73 | 1.26 |
| ACC | 2.481 | 10.3 | 9.7 | 12.7 | 22.8 | 11.7 | 0.24 | 0.26 | 0.20 | 0.27 | 0.21 | |
| ACA | 24.814 | 15.7 | 17.4 | 17.8 | 8.1 | 20.6 | 1.58 | 1.43 | 1.39 | 3.06 | 1.20 | |
| ACG | 9.926 | 7.7 | 4.5 | 8.0 | 15.0 | 3.4 | 1.29 | 2.21 | 1.24 | 0.66 | 2.92 | |
| Ala | GCU | 7.444 | 28.3 | 31.2 | 21.2 | 15.4 | 26.8 | 0.26 | 0.24 | 0.35 | 0.48 | 0.28 |
| GCC | 2.481 | 10.3 | 12.5 | 12.6 | 25.2 | 12.0 | 0.24 | 0.20 | 0.20 | 0.10 | 0.21 | |
| GCA | 22.333 | 17.5 | 23.1 | 16.2 | 20.7 | 23.5 | 1.28 | 0.97 | 1.38 | 1.08 | 0.95 | |
| GCG | 2.481 | 9.0 | 5.8 | 6.2 | 32.3 | 6.9 | 0.28 | 0.43 | 0.40 | 0.08 | 0.36 | |
| Tyr | UAU | 14.888 | 14.6 | 17.8 | 18.8 | 16.5 | 16.5 | 1.02 | 0.84 | 0.79 | 0.90 | 0.90 |
| UAC | 2.481 | 13.7 | 13.5 | 14.8 | 12.3 | 9.7 | 0.18 | 0.18 | 0.17 | 0.20 | 0.26 | |
| His | CAU | 27.295 | 13.8 | 13.4 | 13.6 | 12.8 | 13.5 | 1.98 | 2.04 | 2.01 | 2.13 | 1.51 |
| CAC | 9.926 | 8.7 | 8.7 | 7.8 | 9.4 | 8.3 | 1.14 | 1.14 | 1.27 | 1.06 | 1.20 | |
| Gln | CAA | 27.295 | 19.4 | 20.7 | 27.3 | 14.7 | 18.1 | 1.41 | 1.32 | 1.00 | 1.86 | 1.16 |
| CAG | 22.333 | 15.2 | 15.0 | 12.1 | 29.4 | 16.7 | 1.47 | 1.49 | 1.85 | 0.76 | 1.34 | |
| Asn | AAU | 29.777 | 22.3 | 28.0 | 35.7 | 19.2 | 23.9 | 1.34 | 1.06 | 0.83 | 1.55 | 1.25 |
| AAC | 24.814 | 20.9 | 17.9 | 24.8 | 21.7 | 15.4 | 1.19 | 1.39 | 1.00 | 1.14 | 1.61 | |
| Lys | AAA | 27.295 | 30.8 | 32.6 | 41.9 | 34.0 | 26.4 | 0.89 | 0.84 | 0.65 | 0.80 | 1.03 |
| AAG | 29.777 | 32.7 | 33.5 | 30.8 | 11.0 | 29.5 | 0.91 | 0.89 | 0.97 | 2.71 | 1.01 | |
| Asp | GAU | 54.591 | 36.6 | 36.9 | 37.6 | 32.8 | 42.1 | 1.49 | 1.52 | 1.45 | 1.66 | 1.30 |
| GAC | 14.888 | 17.2 | 16.9 | 20.2 | 19.2 | 14.5 | 0.87 | 0.88 | 0.74 | 0.78 | 1.07 | |
| Glu | GAA | 54.591 | 34.3 | 36.0 | 45.6 | 39.3 | 35.1 | 1.59 | 1.52 | 1.20 | 1.39 | 1.56 |
| GAG | 34.739 | 32.2 | 29.4 | 19.2 | 18.7 | 27.4 | 1.08 | 1.18 | 1.81 | 1.86 | 1.27 | |
| Cys | UGU | 24.814 | 10.5 | 9.8 | 8.1 | 5.2 | 13.4 | 2.36 | 2.53 | 3.06 | 4.77 | 1.85 |
| UGC | 7.444 | 7.2 | 7.2 | 4.8 | 6.4 | 9.4 | 1.03 | 1.03 | 1.55 | 1.16 | 0.79 | |
| Trp | UGG | 14.888 | 12.5 | 12.2 | 10.4 | 15.3 | 11.5 | 1.19 | 1.22 | 1.43 | 0.97 | 1.29 |
| Arg | CGU | 12.407 | 9.0 | 7.5 | 6.4 | 20.2 | 5.7 | 1.38 | 1.65 | 1.94 | 0.61 | 2.18 |
| CGC | 0.000 | 3.8 | 3.9 | 2.6 | 20.8 | 4.0 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| CGA | 4.963 | 6.3 | 5.3 | 3.0 | 3.8 | 4.6 | 0.79 | 0.94 | 1.65 | 1.31 | 1.08 | |
| CGG | 2.481 | 4.9 | 3.7 | 1.7 | 6.2 | 1.8 | 0.51 | 0.67 | 1.46 | 0.40 | 1.38 | |
| Ser | AGU | 7.444 | 14.0 | 13.3 | 14.2 | 9.4 | 14.5 | 0.53 | 0.56 | 0.52 | 0.79 | 0.51 |
| AGC | 13.370 | 11.3 | 10.0 | 9.8 | 16.0 | 10.6 | 1.18 | 1.34 | 1.36 | 0.84 | 1.26 | |
| Arg | AGA | 12.407 | 19.0 | 16.0 | 21.3 | 2.9 | 10.9 | 0.65 | 0.78 | 0.58 | 4.28 | 1.14 |
| AGG | 12.407 | 11.0 | 12.2 | 9.2 | 1.8 | 15.0 | 1.13 | 1.02 | 1.35 | 6.89 | 0.83 | |
| Gly | GGU | 13.370 | 22.2 | 22.3 | 23.9 | 24.2 | 22.9 | 0.06 | 0.60 | 0.56 | 0.55 | 0.58 |
| GGC | 7.444 | 9.2 | 11.2 | 9.8 | 28.1 | 11.6 | 0.81 | 0.66 | 0.76 | 0.26 | 0.64 | |
| GGA | 32.258 | 24.2 | 23.2 | 10.9 | 8.9 | 17.1 | 1.33 | 1.39 | 2.96 | 3.62 | 1.89 | |
| GGG | 22.333 | 10.2 | 10.5 | 6.0 | 11.8 | 14.9 | 2.17 | 2.13 | 3.72 | 1.89 | 1.50 | |
| TER | *UAA | 0.000 | 0.9 | 1.1 | 1.1 | 2.0 | 1.1 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| *UAG | 0.000 | 0.5 | 0.5 | 0.5 | 0.3 | 0.0 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| *UGA | 2.481 | 1.2 | 1.0 | 0.7 | 1.1 | 1.6 | 2.07 | 2.48 | 3.54 | 2.26 | 1.55 |
表4 PmNAC8与部分模式生物基因组密码子使用偏好性比较
Table 4 Comparison of codon usage preference between PmNAC8 and other model organisms
| 氨基酸 Amino acid | 密码子 Codon | 马尾松PmNAC8 | 拟南芥基因组At | 烟草基因组Nt | 酵母基因组Sc | 大肠杆菌基因组Ec | 欧洲山杨基因组Pt | PmNAC8/At | PmNAC8/Nt | PmNAC8/Sc | PmNAC8/Ec | PmNAC8/Pt |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phe | UUU | 24.814 | 21.8 | 25.1 | 26.1 | 22.2 | 28.2 | 1.14 | 0.99 | 0.95 | 1.12 | 0.10 |
| UUC | 7.444 | 20.7 | 18.0 | 18.4 | 15.9 | 17.3 | 0.36 | 0.36 | 0.40 | 0.47 | 0.43 | |
| Leu | UUA | 14.888 | 12.7 | 13.4 | 26.2 | 13.8 | 14.0 | 1.17 | 1.11 | 0.57 | 1.08 | 1.06 |
| UUG | 12.407 | 20.9 | 22.3 | 27.2 | 13.0 | 26.3 | 0.59 | 0.56 | 0.55 | 0.95 | 0.47 | |
| CUU | 17.370 | 24.1 | 24.0 | 12.3 | 11.4 | 22.7 | 0.72 | 0.72 | 1.41 | 1.52 | 0.77 | |
| CUC | 9.926 | 16.1 | 12.3 | 5.4 | 10.5 | 15.6 | 0.62 | 0.81 | 1.84 | 0.95 | 0.64 | |
| CUA | 4.963 | 9.9 | 9.4 | 13.4 | 3.9 | 7.6 | 0.50 | 0.53 | 0.37 | 1.27 | 0.65 | |
| CUG | 0.000 | 9.8 | 10.2 | 10.5 | 51.1 | 8.5 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Ile | AUU | 24.814 | 21.5 | 27.8 | 30.1 | 29.7 | 28.1 | 1.15 | 0.89 | 0.82 | 0.84 | 0.88 |
| AUC | 9.926 | 18.5 | 13.9 | 17.2 | 23.9 | 14.5 | 0.54 | 0.71 | 0.58 | 0.42 | 0.68 | |
| AUA | 17.370 | 12.6 | 14.0 | 17.8 | 5.5 | 15.0 | 1.38 | 1.24 | 0.98 | 3.16 | 1.16 | |
| Met | AUG | 17.370 | 24.5 | 25.0 | 20.9 | 27.2 | 23.6 | 0.71 | 0.69 | 0.83 | 0.64 | 0.74 |
| Val | GUU | 12.407 | 27.2 | 26.8 | 22.1 | 18.1 | 39.1 | 0.46 | 0.46 | 0.56 | 0.69 | 0.32 |
| GUC | 7.444 | 12.8 | 11.1 | 11.8 | 14.8 | 10.8 | 0.58 | 0.67 | 0.63 | 0.50 | 0.69 | |
| GUA | 7.444 | 9.9 | 11.4 | 11.8 | 10.9 | 9.4 | 0.75 | 0.65 | 0.63 | 0.68 | 0.79 | |
| GUG | 12.407 | 17.4 | 16.7 | 10.8 | 26.2 | 19.8 | 0.72 | 0.74 | 1.15 | 0.47 | 0.63 | |
| Ser | UCU | 17.370 | 25.2 | 20.0 | 23.5 | 8.7 | 18.3 | 0.69 | 0.87 | 0.74 | 2.00 | 0.95 |
| UCC | 4.963 | 11.2 | 10.2 | 14.2 | 8.9 | 11.8 | 0.44 | 0.49 | 0.35 | 0.56 | 0.42 | |
| UCA | 32.258 | 18.3 | 17.6 | 18.7 | 8.1 | 21.2 | 1.76 | 1.83 | 1.73 | 3.98 | 1.52 | |
| UCG | 0.000 | 9.3 | 5.3 | 8.6 | 8.8 | 8.3 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Pro | CCU | 27.295 | 18.7 | 18.7 | 13.5 | 7.2 | 15.8 | 1.46 | 1.46 | 2.02 | 3.80 | 1.73 |
| CCC | 9.926 | 5.3 | 6.6 | 6.8 | 5.6 | 8.0 | 1.87 | 1.50 | 1.46 | 1.77 | 1.24 | |
| CCA | 32.258 | 16.1 | 19.8 | 18.3 | 8.4 | 16.1 | 2.00 | 1.63 | 1.76 | 3.84 | 2.00 | |
| CCG | 0.000 | 8.6 | 5.0 | 5.3 | 22.4 | 7.4 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| Thr | ACU | 24.814 | 17.5 | 20.3 | 20.3 | 9.1 | 19.7 | 1.42 | 1.22 | 1.22 | 2.73 | 1.26 |
| ACC | 2.481 | 10.3 | 9.7 | 12.7 | 22.8 | 11.7 | 0.24 | 0.26 | 0.20 | 0.27 | 0.21 | |
| ACA | 24.814 | 15.7 | 17.4 | 17.8 | 8.1 | 20.6 | 1.58 | 1.43 | 1.39 | 3.06 | 1.20 | |
| ACG | 9.926 | 7.7 | 4.5 | 8.0 | 15.0 | 3.4 | 1.29 | 2.21 | 1.24 | 0.66 | 2.92 | |
| Ala | GCU | 7.444 | 28.3 | 31.2 | 21.2 | 15.4 | 26.8 | 0.26 | 0.24 | 0.35 | 0.48 | 0.28 |
| GCC | 2.481 | 10.3 | 12.5 | 12.6 | 25.2 | 12.0 | 0.24 | 0.20 | 0.20 | 0.10 | 0.21 | |
| GCA | 22.333 | 17.5 | 23.1 | 16.2 | 20.7 | 23.5 | 1.28 | 0.97 | 1.38 | 1.08 | 0.95 | |
| GCG | 2.481 | 9.0 | 5.8 | 6.2 | 32.3 | 6.9 | 0.28 | 0.43 | 0.40 | 0.08 | 0.36 | |
| Tyr | UAU | 14.888 | 14.6 | 17.8 | 18.8 | 16.5 | 16.5 | 1.02 | 0.84 | 0.79 | 0.90 | 0.90 |
| UAC | 2.481 | 13.7 | 13.5 | 14.8 | 12.3 | 9.7 | 0.18 | 0.18 | 0.17 | 0.20 | 0.26 | |
| His | CAU | 27.295 | 13.8 | 13.4 | 13.6 | 12.8 | 13.5 | 1.98 | 2.04 | 2.01 | 2.13 | 1.51 |
| CAC | 9.926 | 8.7 | 8.7 | 7.8 | 9.4 | 8.3 | 1.14 | 1.14 | 1.27 | 1.06 | 1.20 | |
| Gln | CAA | 27.295 | 19.4 | 20.7 | 27.3 | 14.7 | 18.1 | 1.41 | 1.32 | 1.00 | 1.86 | 1.16 |
| CAG | 22.333 | 15.2 | 15.0 | 12.1 | 29.4 | 16.7 | 1.47 | 1.49 | 1.85 | 0.76 | 1.34 | |
| Asn | AAU | 29.777 | 22.3 | 28.0 | 35.7 | 19.2 | 23.9 | 1.34 | 1.06 | 0.83 | 1.55 | 1.25 |
| AAC | 24.814 | 20.9 | 17.9 | 24.8 | 21.7 | 15.4 | 1.19 | 1.39 | 1.00 | 1.14 | 1.61 | |
| Lys | AAA | 27.295 | 30.8 | 32.6 | 41.9 | 34.0 | 26.4 | 0.89 | 0.84 | 0.65 | 0.80 | 1.03 |
| AAG | 29.777 | 32.7 | 33.5 | 30.8 | 11.0 | 29.5 | 0.91 | 0.89 | 0.97 | 2.71 | 1.01 | |
| Asp | GAU | 54.591 | 36.6 | 36.9 | 37.6 | 32.8 | 42.1 | 1.49 | 1.52 | 1.45 | 1.66 | 1.30 |
| GAC | 14.888 | 17.2 | 16.9 | 20.2 | 19.2 | 14.5 | 0.87 | 0.88 | 0.74 | 0.78 | 1.07 | |
| Glu | GAA | 54.591 | 34.3 | 36.0 | 45.6 | 39.3 | 35.1 | 1.59 | 1.52 | 1.20 | 1.39 | 1.56 |
| GAG | 34.739 | 32.2 | 29.4 | 19.2 | 18.7 | 27.4 | 1.08 | 1.18 | 1.81 | 1.86 | 1.27 | |
| Cys | UGU | 24.814 | 10.5 | 9.8 | 8.1 | 5.2 | 13.4 | 2.36 | 2.53 | 3.06 | 4.77 | 1.85 |
| UGC | 7.444 | 7.2 | 7.2 | 4.8 | 6.4 | 9.4 | 1.03 | 1.03 | 1.55 | 1.16 | 0.79 | |
| Trp | UGG | 14.888 | 12.5 | 12.2 | 10.4 | 15.3 | 11.5 | 1.19 | 1.22 | 1.43 | 0.97 | 1.29 |
| Arg | CGU | 12.407 | 9.0 | 7.5 | 6.4 | 20.2 | 5.7 | 1.38 | 1.65 | 1.94 | 0.61 | 2.18 |
| CGC | 0.000 | 3.8 | 3.9 | 2.6 | 20.8 | 4.0 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| CGA | 4.963 | 6.3 | 5.3 | 3.0 | 3.8 | 4.6 | 0.79 | 0.94 | 1.65 | 1.31 | 1.08 | |
| CGG | 2.481 | 4.9 | 3.7 | 1.7 | 6.2 | 1.8 | 0.51 | 0.67 | 1.46 | 0.40 | 1.38 | |
| Ser | AGU | 7.444 | 14.0 | 13.3 | 14.2 | 9.4 | 14.5 | 0.53 | 0.56 | 0.52 | 0.79 | 0.51 |
| AGC | 13.370 | 11.3 | 10.0 | 9.8 | 16.0 | 10.6 | 1.18 | 1.34 | 1.36 | 0.84 | 1.26 | |
| Arg | AGA | 12.407 | 19.0 | 16.0 | 21.3 | 2.9 | 10.9 | 0.65 | 0.78 | 0.58 | 4.28 | 1.14 |
| AGG | 12.407 | 11.0 | 12.2 | 9.2 | 1.8 | 15.0 | 1.13 | 1.02 | 1.35 | 6.89 | 0.83 | |
| Gly | GGU | 13.370 | 22.2 | 22.3 | 23.9 | 24.2 | 22.9 | 0.06 | 0.60 | 0.56 | 0.55 | 0.58 |
| GGC | 7.444 | 9.2 | 11.2 | 9.8 | 28.1 | 11.6 | 0.81 | 0.66 | 0.76 | 0.26 | 0.64 | |
| GGA | 32.258 | 24.2 | 23.2 | 10.9 | 8.9 | 17.1 | 1.33 | 1.39 | 2.96 | 3.62 | 1.89 | |
| GGG | 22.333 | 10.2 | 10.5 | 6.0 | 11.8 | 14.9 | 2.17 | 2.13 | 3.72 | 1.89 | 1.50 | |
| TER | *UAA | 0.000 | 0.9 | 1.1 | 1.1 | 2.0 | 1.1 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| *UAG | 0.000 | 0.5 | 0.5 | 0.5 | 0.3 | 0.0 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| *UGA | 2.481 | 1.2 | 1.0 | 0.7 | 1.1 | 1.6 | 2.07 | 2.48 | 3.54 | 2.26 | 1.55 |

图7 马尾松PmNAC8在不同组织中的表达水平 F:包含花粉的完整花;NL:当年生新叶;OL:当年生老叶;NS:当年生未成熟茎;OS:当年生成熟茎;R:根;M:木质部;P:韧皮部。幼叶组织为对照,同一列中不同小写字母表示差异显著,P<0.05。下同
Fig.7 PmNAC8 expression level in the different tissues of P. massoniana F:Whole flowers containing pollen. NL:New leaves of the current year. OL:Old leaves. NS:Immature stem of current year. OS:Mature stem current year. R:Root. M:Xylem. P:Phloem. The young leaf tissue is used as a control. Different lowercase letters in the same column indicate a significant difference,P<0.05. The same below
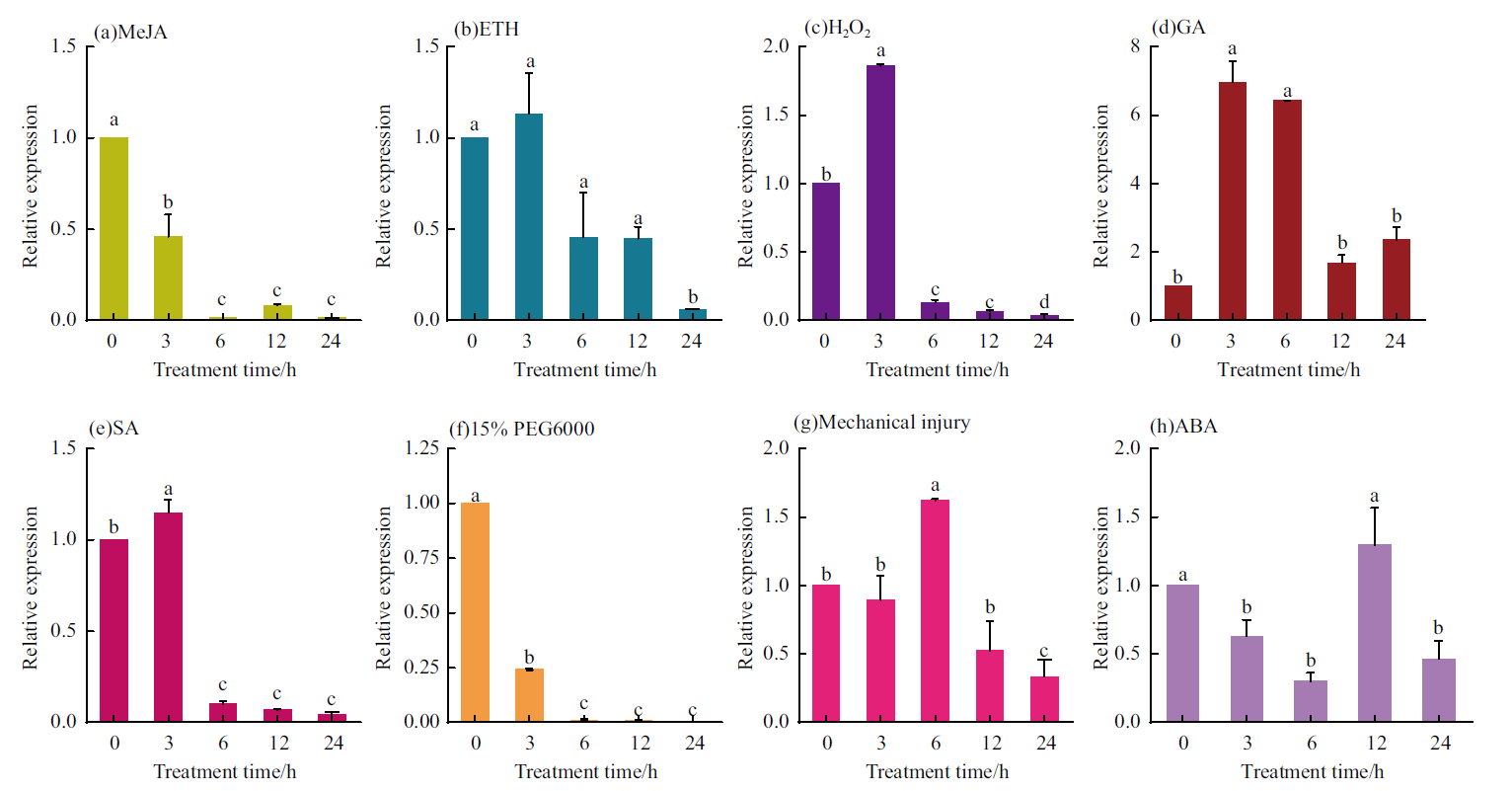
图8 马尾松PmNAC8在不同胁迫下的表达水平 以上所有处理均以0 h作为对照;(a):10 mmol/L MeJA;(b):50 μmol/L ETH;(c):10 mmol/L H2O2;(d):2 mmol/L GA;(e):1 mmol/L SA;(f):15% PEG6000;(g):机械损伤胁迫;(h):400 μmol/L ABA
Fig.8 PmNAC8 expression of P. massonniana under different stress All the above treatments are treated with 0 h as control.(a):10 mmol/L MeJA;(b):50 μmol/L ETHs;(c):10 mmol/L H2O2;(d):2 mmol/L GA;(e):1 mmol/L SA;(f):15% PEG6000;(g):mechanical injury stress;(h):400 μmol/L ABA
图9 PmNAC8启动子扩增电泳 M:DL2502 DAN marker,1-3泳道为PmNAC8启动子PCR扩增
Fig. 9 Amplification electrophoresis of PmNAC8 promoter M:DL2502 DNA marker,1-3:PCR amplifification of PmNAC8 promoter
| 元件Element | 功能Function | 核心序列Core sequence | 数量Amount |
|---|---|---|---|
| Box 4 | 参与光响应部分保守DNA组件 Part of a conserved DNA module involved in light responsiveness | ATTAAT | 2 |
| G-Box | 参与光响应的顺式调控元件 Cis-acting regulatory element involved in light responsiveness | CACGAC | 2 |
| GT1-motif | 光响应元件Light responsive element | GGTTAAT | 1 |
| MRE | 涉及光反应的MYB结合位点 MYB binding sites involved in light reactions | AACCTAA | 1 |
| MBS | 参与干旱诱导MYB结合位点 MYB binding site involved in drought induction | CAACTG | 1 |
| ARE | 抗氧化反应元件 Antioxidant reaction element | AAACCA | 2 |
| O2-site | 参与玉米醇溶蛋白的代谢调节 Cis-acting regulatory element involved in zein metabolism regulation | GTTGACGTGA | 1 |
| TATA-box | 转录起始-30处核心启动元件 Core promoter element around -30 of transcription start | TACAAAA | 21 |
| HD-Zip 3 | 蛋白结合位点 Protein binding site | GTAAT(G/C)ATTAC | 3 |
| CAAT-box | 启动子和增强子区域共有顺式作用元件 Common cis-acting element in promoter and enhancer regions | CAAT/CAAAT/CCAAT/TGCCAAC/ | 34 |
| P-box | 赤霉素响应顺式作用元件 Cis-acting element involved in gibberellin responsiveness | CCTTTTG | 1 |
表5 PmNAC8启动子中的顺式作用元件
Table 5 Cis-acting elements in the PmNAC8 promoter
| 元件Element | 功能Function | 核心序列Core sequence | 数量Amount |
|---|---|---|---|
| Box 4 | 参与光响应部分保守DNA组件 Part of a conserved DNA module involved in light responsiveness | ATTAAT | 2 |
| G-Box | 参与光响应的顺式调控元件 Cis-acting regulatory element involved in light responsiveness | CACGAC | 2 |
| GT1-motif | 光响应元件Light responsive element | GGTTAAT | 1 |
| MRE | 涉及光反应的MYB结合位点 MYB binding sites involved in light reactions | AACCTAA | 1 |
| MBS | 参与干旱诱导MYB结合位点 MYB binding site involved in drought induction | CAACTG | 1 |
| ARE | 抗氧化反应元件 Antioxidant reaction element | AAACCA | 2 |
| O2-site | 参与玉米醇溶蛋白的代谢调节 Cis-acting regulatory element involved in zein metabolism regulation | GTTGACGTGA | 1 |
| TATA-box | 转录起始-30处核心启动元件 Core promoter element around -30 of transcription start | TACAAAA | 21 |
| HD-Zip 3 | 蛋白结合位点 Protein binding site | GTAAT(G/C)ATTAC | 3 |
| CAAT-box | 启动子和增强子区域共有顺式作用元件 Common cis-acting element in promoter and enhancer regions | CAAT/CAAAT/CCAAT/TGCCAAC/ | 34 |
| P-box | 赤霉素响应顺式作用元件 Cis-acting element involved in gibberellin responsiveness | CCTTTTG | 1 |
| [1] |
Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses[J]. Curr Opin Plant Biol, 2003, 6(5):410-417.
pmid: 12972040 |
| [2] |
Rejeb IB, Pastor V, Mauch-Mani B. Plant responses to simultaneous biotic and abiotic stress:molecular mechanisms[J]. Plants-Basel, 2014, 3(4):458-475.
doi: 10.3390/plants3040458 pmid: 27135514 |
| [3] |
Liu H, Zhou Y, Li H, et al. Molecular and functional characterization of ShNAC1, an NAC transcription factor from Solanum habrochaites[J]. Plant Sci, 2018, 271:9-19.
doi: 10.1016/j.plantsci.2018.03.005 URL |
| [4] |
Long L, et al. GbMPK3, a mitogen-activated protein kinase from cotton, enhances drought and oxidative stress tolerance in tobacco[J]. Plant Cell Tissue Organ Cult, 2014, 116(2):153-162.
doi: 10.1007/s11240-013-0392-1 URL |
| [5] | Shao H, Wang H, Tang X. NAC transcription factors in plant multiple abiotic stress responses:progress and prospects[J]. Front Plant Sci, 2015, 6:902. |
| [6] |
Duval M, Hsieh TF, Kim SY, et al. Molecular characterization of AtNAM:a member of the Arabidopsis NAC domain superfamily[J]. Plant Mol Biol, 2002, 50(2):237-248.
doi: 10.1023/A:1016028530943 URL |
| [7] |
Tran LS, et al. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive Cis-element in the early responsive to dehydration stress 1 promoter[J]. Plant Cell, 2004, 16(9):2481-2498.
doi: 10.1105/tpc.104.022699 URL |
| [8] |
Chen X, Wang Y, Lv B, et al. The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway[J]. Plant Cell Physiol, 2014, 55(3):604-619.
doi: 10.1093/pcp/pct204 pmid: 24399239 |
| [9] |
Xue GP, Way HM, et al. Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat[J]. Mol Plant, 2011, 4(4):697-712.
doi: 10.1093/mp/ssr013 URL |
| [10] |
Lu M, Ying S, Zhang DF, et al. A maize stress-responsive NAC transcription factor, ZmSNAC1, confers enhanced tolerance to dehydration in transgenic Arabidopsis[J]. Plant Cell Rep, 2012, 31(9):1701-1711.
doi: 10.1007/s00299-012-1284-2 URL |
| [11] |
Puranik S, Sahu PP, et al. NAC proteins:regulation and role in stress tolerance[J]. Trends Plant Sci, 2012, 17(6):369-381.
doi: 10.1016/j.tplants.2012.02.004 pmid: 22445067 |
| [12] |
Mahmood K, El-Kereamy A, Kim SH, et al. ANAC032 positively regulates age-dependent and stress-induced senescence in Arabidopsis thaliana[J]. Plant Cell Physiol, 2016, 57(10):2029-2046.
pmid: 27388337 |
| [13] |
Mahmood K, Xu Z, El-Kereamy A, et al. The Arabidopsis transcription factor ANAC032 represses anthocyanin biosynjournal in response to high sucrose and oxidative and abiotic stresses[J]. Front Plant Sci, 2016, 7:1548.
pmid: 27790239 |
| [14] |
Oda-Yamamizo C, Mitsuda N, Sakamoto S, et al. The NAC transcription factor ANAC046 is a positive regulator of chlorophyll degradation and senescence in Arabidopsis leaves[J]. Sci Rep, 2016, 6:23609.
doi: 10.1038/srep23609 pmid: 27021284 |
| [15] | 蒙露露, 何冠谛, 田维军, 等. 马铃薯StNAC2基因的克隆及其在镉胁迫下的表达分析[J]. 分子植物育种, 2020, 18(22):7293-7300. |
| Meng LL, He GD, Tian WJ, et al. Cloning and expression analysis of StNAC2 gene in potato(Solanum tuberosum L.)under cadmium stress[J]. Mol Plant Breed, 2020, 18(22):7293-7300. | |
| [16] |
Jeong JS, Kim YS, Redillas MCFR, et al. OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field[J]. Plant Biotechnol J, 2013, 11(1):101-114.
doi: 10.1111/pbi.12011 pmid: 23094910 |
| [17] | 岳文冉, 岳俊燕, 张秀娟, 等. 中间锦鸡儿CiNAC1基因促进转基因拟南芥叶片的衰老[J]. 中国生物工程杂志, 2018, 38(4):24-29. |
| Yue WR, Yue JY, Zhang XJ, et al. The CiNAC1 from Caragana intermedia promotes transgenic Arabidopsis leaf senescence[J]. China Biotechnol, 2018, 38(4):24-29. | |
| [18] |
Fang L, Su L, Sun X, et al. Expression of Vitis amurensis NAC26 in Arabidopsis enhances drought tolerance by modulating jasmonic acid synjournal[J]. J Exp Bot, 2016, 67(9):2829-2845.
doi: 10.1093/jxb/erw122 URL |
| [19] |
武肖琦, 冯涛, 刘艳军, 等. 桃PpNAC72的克隆及表达分析[J]. 生物技术通报, 2019, 35(6):16-23.
doi: 10.13560/j.cnki.biotech.bull.1985.2018-0891 |
| Wu XQ, Feng T, et al. Cloning and expression analysis of the PpNAC72 in peach[J]. Biotechnol Bull, 2019, 35(6):16-23. | |
| [20] | 徐向华, 丁贵杰. 马尾松适应低磷胁迫的生理生化响应[J]. 林业科学, 2006, 42(9):24-28. |
| Xu XH, Ding GJ. Physiological and biochemical responses of Pinus massoniana to low phosphorus stress[J]. Sci Silvae Sin, 2006, 42(9):24-28. | |
| [21] | 王晓锋, 何卫龙, 蔡卫佳, 等. 马尾松转录组测序和分析[J]. 分子植物育种, 2013, 11(3):385-392. |
| Wang XF, He WL, Cai WJ, et al. Analysis on transcriptome sequenced for Pinus massoniana[J]. Mol Plant Breed, 2013, 11(3):385-392. | |
| [22] |
Kumar S, Stecher G, Li M, et al. MEGA X:molecular evolutionary genetics analysis across computing platforms[J]. Mol Biol Evol, 2018, 35(6):1547-1549.
doi: 10.1093/molbev/msy096 URL |
| [23] | 赵春丽, 彭丽云, 王晓, 等. 苋菜AtGAI基因密码子偏好性与进化分析[J]. 中国农业大学学报, 2019, 24(12):10-22. |
| Zhao CL, Peng LY, Wang X, et al. Codon bias and evolution analysis of AtGAI in Amaranthus tricolor L[J]. J China Agric Univ, 2019, 24(12):10-22. | |
| [24] | 薛拥志, 王宝地, 王连荣. 杏扁PaCBF1基因密码子偏好性分析[J]. 分子植物育种, 2018, 16(15):4890-4898. |
| Xue YZ, Wang BD, Wang LR. Analysis of codon bias of PaCBF1 gene in Prunus armeniaca L[J]. Mol Plant Breed, 2018, 16(15):4890-4898. | |
| [25] | 李尊强, 王春军, 杨爱国, 等. 烟草DXS基因的克隆及其亚细胞定位分析[J]. 安徽农业科学, 2013, 41(30):11957-11960. |
| Li ZQ, Wang CJ, Yang AG, et al. Cloning and subcellular localization of DXS gene in tobacco[J]. J Anhui Agric Sci, 2013, 41(30):11957-11960. | |
| [26] |
Yao S, Wu F, Hao QQ, et al. Transcriptome-wide identification of WRKY transcription factors and their expression profiles under different types of biological and abiotic stress in Pinus massoniana lamb[J]. Genes, 2020, 11(11):1386.
doi: 10.3390/genes11111386 URL |
| [27] |
Zhu PH, Ma YY, Zhu LZ, et al. Selection of suitable reference genes in Pinus massoniana lamb. under different abiotic stresses for qPCR normalization[J]. Forests, 2019, 10(8):632.
doi: 10.3390/f10080632 URL |
| [28] | 朱灵芝, 朱沛煌, 李荣, 等. 马尾松PmDXR基因密码子偏好性分析[J]. 林业科学研究, 2021, 34(2):102-113. |
| Zhu LZ, Zhu PH, Li R, et al. Analysis on codon bias of PmDXR gene in Pinus massoniana lamb[J]. For Res, 2021, 34(2):102-113. | |
| [29] | Ondati Evans. 陆地棉GhNAC18和GhNAC20基因在叶片衰老和胁迫应答中的功能分析[D]. 北京:中国农业科学院, 2016. |
| Ondati E. Functional analysis of GhNAC18 and GhNAC20 during leaf senescence and stress responses in cotton(Gossypium hirsutum L.)[D]. Beijing:Chinese Academy of Agricultural Sciences, 2016. | |
| [30] |
Jiang G, Yan H, Wu F, et al. Litchi fruit LcNAC1 is a target of LcMYC2 and regulator of fruit senescence through its interaction with LcWRKY1[J]. Plant Cell Physiol, 2017, 58(6):1075-1089.
doi: 10.1093/pcp/pcx054 URL |
| [31] | 张海娟, 吴剑锋, 胡帅, 等. 芜菁NAC转录因子BcNAC2基因的分离及其表达[J]. 园艺学报, 2011, 38(6):1089-1096. |
| Zhang HJ, Wu JF, Hu S, et al. Isolation and expression analysis of BcNAC2, a NAC transcription factor gene in turnip[J]. Acta Hortic Sin, 2011, 38(6):1089-1096. | |
| [32] |
Yang X, Hu YX, Li XL, et al. Molecular characterization and function analysis of SlNAC2 in Suaeda liaotungensis K[J]. Gene, 2014, 543(2):190-197.
doi: 10.1016/j.gene.2014.04.025 URL |
| [33] |
Yang SD, Seo PJ, Yoon HK, et al. The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes[J]. Plant Cell, 2011, 23(6):2155-2168.
doi: 10.1105/tpc.111.084913 URL |
| [34] |
荣玉萍, 唐彬, 李鹏, 等. 苦荞NAC转录因子FtNAC17的鉴定及表达分析[J]. 生物技术通报, 2021, 37(1):174-181.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-0556 |
| Rong YP, Tang B, Li P, et al. Identification and expression of NAC transcription factor FtNAC17 in Tartary buckwheat[J]. Biotechnol Bull, 2021, 37(1):174-181. | |
| [35] | 孙欣, 上官凌飞, 房经贵, 等. 葡萄NAC转录因子家族生物信息学分析[J]. 基因组学与应用生物学, 2011, 30(2):229-242. |
| Sun X, Shangguan LF, Fang JG, et al. Bioinformatics analysis of the NAC transcription factor family in grapevine[J]. Genom Appl Biol, 2011, 30(2):229-242. | |
| [36] | 朱沛煌, 陈妤, 朱灵芝, 等. 马尾松转录组密码子使用偏好性及其影响因素[J]. 林业科学, 2020, 56(4):74-81. |
| Zhu PH, Chen Y, Zhu LZ, et al. Codon usage bias and its influencing factors in Pinus massoniana transcriptome[J]. Sci Silvae Sin, 2020, 56(4):74-81. | |
| [37] | 杨晓娜, 赵昶灵, 李云, 等. 启动子序列克隆和功能分析方法的研究进展[J]. 云南农业大学学报:自然科学版, 2010, 25(2):283-290. |
| Yang XN, Zhao CL, Li Y, et al. Research advances in the methods of cloning and function-analyzing of promoters[J]. J Yunnan Agric Univ:Nat Sci, 2010, 25(2):283-290. | |
| [38] | 颜静宛, 林智敏, 周淑芬, 等. 水稻胚特异表达基因Os08PTS启动子的克隆及分析[J]. 福建农业科技, 2021, 52(3):6-10. |
| Yan JW, Lin ZM, Zhou SF, et al. Cloning and analysis of the promoter of rice embryo-specific expression gene Os08PTS[J]. Fujian Agric Sci Technol, 2021, 52(3):6-10. | |
| [39] |
Puranik S, Bahadur RP, Srivastava PS, et al. Molecular cloning and characterization of a membrane associated NAC family gene, SiNAC from foxtail millet[Setaria italica(L.)P. Beauv][J]. Mol Biotechnol, 2011, 49(2):138-150.
doi: 10.1007/s12033-011-9385-7 pmid: 21312005 |
| [1] | 刘玉玲, 王梦瑶, 孙琦, 马利花, 朱新霞. 启动子RD29A对转雪莲SikCDPK1基因烟草抗逆性的影响[J]. 生物技术通报, 2023, 39(9): 168-175. |
| [2] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [3] | 李博, 刘合霞, 陈宇玲, 周兴文, 朱宇林. 金花茶CnbHLH79转录因子的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 241-250. |
| [4] | 赵雪婷, 高利燕, 王俊刚, 沈庆庆, 张树珍, 李富生. 甘蔗AP2/ERF转录因子基因ShERF3的克隆、表达及其编码蛋白的定位[J]. 生物技术通报, 2023, 39(6): 208-216. |
| [5] | 郭怡婷, 赵文菊, 任延靖, 赵孟良. 菊芋NAC转录因子家族基因的鉴定及分析[J]. 生物技术通报, 2023, 39(6): 217-232. |
| [6] | 徐红云, 吕俊, 于存. 根际溶磷伯克霍尔德菌Paraburkholderia spp.对马尾松苗的促生作用[J]. 生物技术通报, 2023, 39(6): 274-285. |
| [7] | 李苑虹, 郭昱昊, 曹燕, 祝振洲, 王飞飞. 外源植物激素调控微藻生长及目标产物积累研究进展[J]. 生物技术通报, 2023, 39(6): 61-72. |
| [8] | 冯珊珊, 王璐, 周益, 王幼平, 方玉洁. WOX家族基因调控植物生长发育和非生物胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(5): 1-13. |
| [9] | 翟莹, 李铭杨, 张军, 赵旭, 于海伟, 李珊珊, 赵艳, 张梅娟, 孙天国. 异源表达大豆转录因子GmNF-YA19提高转基因烟草抗旱性[J]. 生物技术通报, 2023, 39(5): 224-232. |
| [10] | 姚姿婷, 曹雪颖, 肖雪, 李瑞芳, 韦小妹, 邹承武, 朱桂宁. 火龙果溃疡病菌实时荧光定量PCR内参基因的筛选[J]. 生物技术通报, 2023, 39(5): 92-102. |
| [11] | 郭三保, 宋美玲, 李灵心, 尧子钊, 桂明明, 黄胜和. 斑地锦查尔酮合酶基因及启动子的克隆与分析[J]. 生物技术通报, 2023, 39(4): 148-156. |
| [12] | 杨岚, 张晨曦, 樊学伟, 王阳光, 王春秀, 李文婷. 鸡 BMP15 基因克隆、表达模式及启动子活性分析[J]. 生物技术通报, 2023, 39(4): 304-312. |
| [13] | 杨春洪, 董璐, 陈林, 宋丽. 大豆VAS1基因家族的鉴定及参与侧根发育的研究[J]. 生物技术通报, 2023, 39(3): 133-142. |
| [14] | 苗淑楠, 高宇, 李昕儒, 蔡桂萍, 张飞, 薛金爱, 季春丽, 李润植. 大豆GmPDAT1参与油脂合成和非生物胁迫应答的功能分析[J]. 生物技术通报, 2023, 39(2): 96-106. |
| [15] | 许睿, 祝英方. 中介体复合物在植物非生物胁迫应答中的功能[J]. 生物技术通报, 2023, 39(11): 54-60. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||