








生物技术通报 ›› 2022, Vol. 38 ›› Issue (5): 240-247.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1183
周子琦1( ), 张洋子1, 兰欣悦1, 刘洋儿2, 朱龙佼2, 许文涛2(
), 张洋子1, 兰欣悦1, 刘洋儿2, 朱龙佼2, 许文涛2( )
)
收稿日期:2021-09-14
出版日期:2022-05-26
发布日期:2022-06-10
作者简介:周子琦,女,硕士研究生,研究方向:适配体的筛选及病原菌检测;E-mail: 基金资助:
ZHOU Zi-qi1( ), ZHANG Yang-zi1, LAN Xin-yue1, LIU Yang-er2, ZHU Long-jiao2, XU Wen-tao2(
), ZHANG Yang-zi1, LAN Xin-yue1, LIU Yang-er2, ZHU Long-jiao2, XU Wen-tao2( )
)
Received:2021-09-14
Published:2022-05-26
Online:2022-06-10
摘要:
发光核酸适配体(light-up nucleic acid aptamers,LNAs)是一类能够特异性结合目标分子并增强目标分子发光性能的功能核酸(functional nucleic acids,FNAs)。LNAs具有组成简单、结构稳定、表达时间短等优点,因此在细胞成像、物质检测等传感领域显示出巨大潜力。随着对细胞内基因原位分析的研究深入,现存胞内成像技术的不足逐渐显露,LNAs成像系统应运而生。本文梳理并概括了不同类型LNAs的筛选方法,并从胞内、体外两个方面对LNAs的前沿应用进行了分析,指出了当前LNAs研究以及传感系统存在的不足,展望了LNAs在无细胞传感中的巨大发展前景,以期能够促进LNAs系统成为胞内外传感研究的一件利器。
周子琦, 张洋子, 兰欣悦, 刘洋儿, 朱龙佼, 许文涛. 发光核酸适配体的筛选及应用[J]. 生物技术通报, 2022, 38(5): 240-247.
ZHOU Zi-qi, ZHANG Yang-zi, LAN Xin-yue, LIU Yang-er, ZHU Long-jiao, XU Wen-tao. Selection and Application of Light-up Nucleic Acid Aptamers[J]. Biotechnology Bulletin, 2022, 38(5): 240-247.
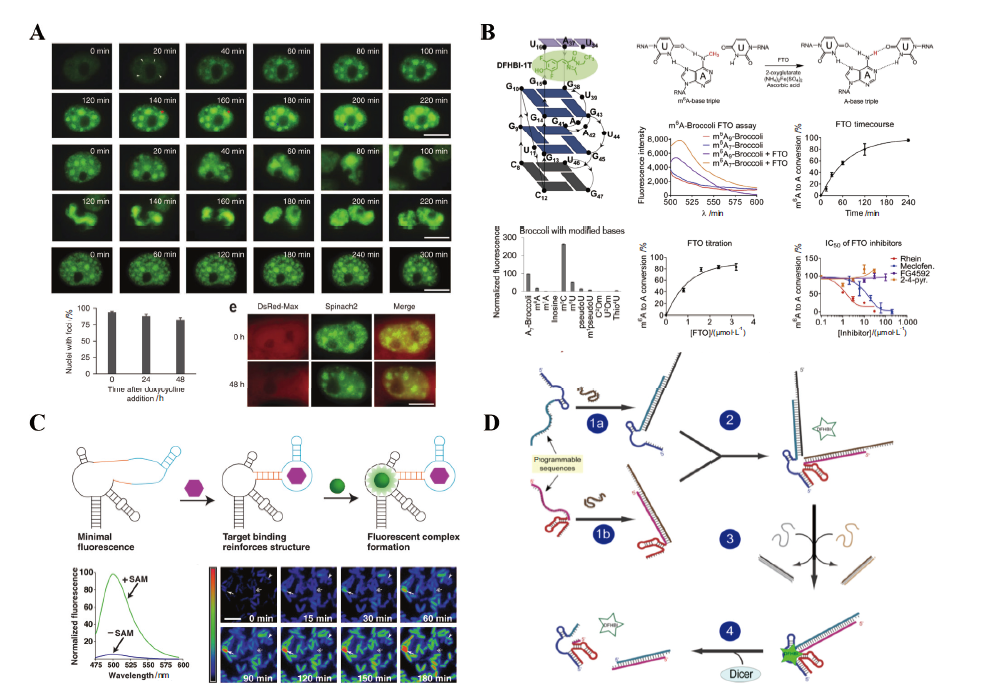
图2 LNAs系统在活细胞内的应用 A:瞬时转染COS-7细胞中(CGG)60-Spinach 2病灶形成的监测;B:m6A修饰Broccoli作为荧光底物在FTO检测中的应用;C:在活细胞中使用RNA适配体和Spinach发光核酸适配体检测SAM;D:劈裂Spinach发光核酸适配体的组装与传感
Fig. 2 Application of LNAs system in living cells A: Monitoring the formation of (CGG)60-Spinach 2 foci in transiently transfected COS-7 cell. B: Application of m6A-modified Broccoli as a fluorometric substrate in the FTO assay. C: Imaging SAM in living cells with RNA aptamer and Spinach. D: Assembly and sensing of aptamer in split Spinach

图3 LNAs系统在体外的应用 A:基于核糖开关的荧光二价金属离子传感器示意图,czcD-2的二级结构和厌氧滴定实验;B:蔬菜适配体标记的RNA及其裂解产物的凝胶检测示意图;C:ROSALIND系统的传感设备与原理
Fig. 3 Application of LNAs system in vitro A: Schematic diagram of fluorescent bivalent metal ion sensor based on riboswitch, secondary structure of czcD-2 and anaerobic titration experiment. B: Schematic representation of the in-gel detection of vegetable aptamer-tagged RNAs and their cleavage products. C: Sensing equipment and principle of ROSALIND system
| [1] | Valeur B. Molecular fluorescence[J]. Digital Encyclopedia of Applied Physics, 2003:477-531. |
| [2] |
Urbanek MO, Galka-Marciniak P, Olejniczak M, et al. RNA imaging in living cells - methods and applications[J]. RNA Biol, 2014, 11(8):1083-1095.
doi: 10.4161/rna.35506 URL |
| [3] |
Wang YX, Shyy JYJ, Chien S. Fluorescence proteins, live-cell imaging, and mechanobiology:seeing is believing[J]. Annu Rev Biomed Eng, 2008, 10:1-38.
doi: 10.1146/annurev.bioeng.010308.161731 URL |
| [4] |
Giepmans BNG, Adams SR, Ellisman MH, et al. The fluorescent toolbox for assessing protein location and function[J]. Science, 2006, 312(5771):217-224.
doi: 10.1126/science.1124618 pmid: 16614209 |
| [5] |
Schnell U, Dijk F, Sjollema KA, et al. Immunolabeling artifacts and the need for live-cell imaging[J]. Nat Methods, 2012, 9(2):152-158.
doi: 10.1038/nmeth.1855 pmid: 22290187 |
| [6] |
Santangelo PJ. Molecular beacons and related probes for intracellular RNA imaging[J]. WIREs Nanomed Nanobiotechnol, 2010, 2(1):11-19.
doi: 10.1002/wnan.52 URL |
| [7] | Ouellet J. RNA fluorescence with light-up aptamers[J]. Front Chem, 2016, 4:29. |
| [8] |
Fusco D, Accornero N, Lavoie B, et al. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells[J]. Curr Biol, 2003, 13(2):161-167.
doi: 10.1016/S0960-9822(02)01436-7 URL |
| [9] |
Babendure JR, Adams SR, Tsien RY. Aptamers switch on fluorescence of triphenylmethane dyes[J]. J Am Chem Soc, 2003, 125(48):14716-14717.
pmid: 14640641 |
| [10] |
Bouhedda F, Fam KT, Collot M, et al. A dimerization-based fluorogenic dye-aptamer module for RNA imaging in live cells[J]. Nat Chem Biol, 2020, 16(1):69-76.
doi: 10.1038/s41589-019-0381-8 pmid: 31636432 |
| [11] |
Chen X, Zhang D, Su N, et al. Visualizing RNA dynamics in live cells with bright and stable fluorescent RNAs[J]. Nat Biotechnol, 2019, 37(11):1287-1293.
doi: 10.1038/s41587-019-0249-1 URL |
| [12] |
Gao T, Luo Y, Li W, et al. Progress in the isolation of aptamers to light-up the dyes and the applications[J]. Analyst, 2020, 145(3):701-718.
doi: 10.1039/C9AN01825E URL |
| [13] |
Swetha P, Fan Z, Wang FL, et al. Genetically encoded light-up RNA aptamers and their applications for imaging and biosensing[J]. J Mater Chem B, 2020, 8(16):3382-3392.
doi: 10.1039/C9TB02668A URL |
| [14] |
Stoltenburg R, Reinemann C, Strehlitz B. SELEX——a(r)evolutionary method to generate high-affinity nucleic acid ligands[J]. Biomol Eng, 2007, 24(4):381-403.
pmid: 17627883 |
| [15] |
Islam MM, Ghielmetti VM, Allen PB. Graphene oxide assisted light-up aptamer selection against Thioflavin T for label-free detection of microRNA[J]. Sci Rep, 2021, 11(1):4291.
doi: 10.1038/s41598-021-83640-z URL |
| [16] |
Chen Y, Wang J, Zhang Y, et al. Selection and characterization of a DNA aptamer to crystal violet[J]. Photochem Photobiol Sci, 2018, 17(6):800-806.
doi: 10.1039/C7PP00457E URL |
| [17] | Sando S, Narita A, Hayami M, et al. Transcription monitoring using fused RNA with a dye-binding light-up aptamer as a tag:a blue fluorescent RNA[J]. Chem Commun:Camb, 2008(33):3858-3860. |
| [18] |
Sando S, Narita A, Aoyama Y. Light-up Hoechst-DNA aptamer pair:generation of an aptamer-selective fluorophore from a conventional DNA-staining dye[J]. Chembiochem, 2007, 8(15):1795-1803.
doi: 10.1002/cbic.200700325 URL |
| [19] |
Wang HY, Wang JN, Sun N, et al. Selection and characterization of malachite green aptamers for the development of light-up probes[J]. ChemistrySelect, 2016, 1(8):1571-1574.
doi: 10.1002/slct.201600154 URL |
| [20] |
Wang JN, Zhang YJ, Wang HY, et al. Selection and analysis of DNA aptamers to berberine to develop a label-free light-up fluorescent probe[J]. New J Chem, 2016, 40(11):9768-9773.
doi: 10.1039/C6NJ02290A URL |
| [21] |
Wang HY, Wang JN, Xu LJ, et al. Selection and characterization of thioflavin T aptamers for the development of light-up probes[J]. Anal Methods, 2016, 8(48):8461-8465.
doi: 10.1039/C6AY02890J URL |
| [22] |
Kaur H. Recent developments in cell-SELEX technology for aptamer selection[J]. Biochim Biophys Acta Gen Subj, 2018, 1862(10):2323-2329.
doi: 10.1016/j.bbagen.2018.07.029 URL |
| [23] |
Paige JS, Wu KY, Jaffrey SR. RNA mimics of green fluorescent protein[J]. Science, 2011, 333(6042):642-646.
doi: 10.1126/science.1207339 URL |
| [24] |
Wang H, Wang J, Wang Q, et al. Selection and characterization of dimethylindole red DNA aptamers for the development of light-up fluorescent probes[J]. Talanta, 2017, 168:217-221.
doi: 10.1016/j.talanta.2017.03.041 URL |
| [25] |
Zou J, Huang X, Wu L, et al. Selection of intracellularly functional RNA mimics of green fluorescent protein using fluorescence-activated cell sorting[J]. J Mol Evol, 2015, 81(5/6):172-178.
doi: 10.1007/s00239-015-9718-4 URL |
| [26] |
Duchardt-Ferner E, Juen M, Bourgeois B, et al. Structure of an RNA aptamer in complex with the fluorophore tetramethylrhodamine[J]. Nucleic Acids Res, 2020, 48(2):949-961.
doi: 10.1093/nar/gkz1113 pmid: 31754719 |
| [27] |
Filonov GS, Moon JD, Svensen N, et al. Broccoli:rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution[J]. J Am Chem Soc, 2014, 136(46):16299-16308.
doi: 10.1021/ja508478x URL |
| [28] |
Lee J, Lee KH, Jeon J, et al. Combining SELEX screening and rational design to develop light-up fluorophore-RNA aptamer pairs for RNA tagging[J]. ACS Chem Biol, 2010, 5(11):1065-1074.
doi: 10.1021/cb1001894 URL |
| [29] |
Ryckelynck M, Baudrey S, Rick C, et al. Using droplet-based microfluidics to improve the catalytic properties of RNA under multiple-turnover conditions[J]. RNA, 2015, 21(3):458-469.
doi: 10.1261/rna.048033.114 pmid: 25605963 |
| [30] |
Autour A, Westhof E, Ryckelynck M. iSpinach:a fluorogenic RNA aptamer optimized for in vitro applications[J]. Nucleic Acids Res, 2016, 44(6):2491-2500.
doi: 10.1093/nar/gkw083 URL |
| [31] |
Armstrong-Price DE, Deore PS, Manderville RA. Intrinsic “turn-on” aptasensor detection of ochratoxin A using energy-transfer fluorescence[J]. J Agric Food Chem, 2020, 68(7):2249-2255.
doi: 10.1021/acs.jafc.9b07391 URL |
| [32] |
Dolgosheina EV, Jeng SC, Panchapakesan SS, et al. RNA mango aptamer-fluorophore:a bright, high-affinity complex for RNA labeling and tracking[J]. ACS Chem Biol, 2014, 9(10):2412-2420.
doi: 10.1021/cb500499x pmid: 25101481 |
| [33] | Okuda M, Fourmy D, Yoshizawa S. Use of Baby Spinach and Broccoli for imaging of structured cellular RNAs[J]. Nucleic Acids Res, 2017, 45(3):1404-1415. |
| [34] |
Strack RL, Disney MD, Jaffrey SR. A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat-containing RNA[J]. Nat Methods, 2013, 10(12):1219-1224.
doi: 10.1038/nmeth.2701 pmid: 24162923 |
| [35] |
Gao S, Zhang S, Sun X, et al. Fluorescent aptasensor based on G-quadruplex-assisted structural transformation for the detection of biomarker lipocalin 1[J]. Biosens Bioelectron, 2020, 169:112607.
doi: 10.1016/j.bios.2020.112607 URL |
| [36] |
Nilaratanakul V, Hauer DA, Griffin DE. Development of encoded Broccoli RNA aptamers for live cell imaging of Alphavirus genomic and subgenomic RNAs[J]. Sci Rep, 2020, 10(1):5233.
doi: 10.1038/s41598-020-61573-3 pmid: 32251299 |
| [37] | Renuka RM, Maroli N, Achuth J, et al. Fret based aptamer assay for sensitive detection of salmonella paratyphi a and revealing its molecular interaction with DNA gyrase[J]. bioRxiv, 2020. DOI: https://doi.org/10.1101/2020.04.02.021881. |
| [38] |
Shi L, Peng P, Zheng J, et al. I-Motif/miniduplex hybrid structures bind benzothiazole dyes with unprecedented efficiencies:a generic light-up system for label-free DNA nanoassemblies and bioimaging[J]. Nucleic Acids Res, 2020, 48(4):1681-1690.
doi: 10.1093/nar/gkaa020 URL |
| [39] |
Yoon T, Kim S, Shin J, et al. Highly sensitive multiplex detection of microRNA using light-up RNA aptamers[J]. Sens Actuat B:Chem, 2021, 330:129410.
doi: 10.1016/j.snb.2020.129410 URL |
| [40] |
Zhang J, Wang L, Jäschke A, et al. A color-shifting near-infrared fluorescent aptamer-fluorophore module for live-cell RNA imaging[J]. Angew Chem Int Ed Engl, 2021, 60(39):21441-21448.
doi: 10.1002/anie.202107250 URL |
| [41] |
Zhou YY, Zuo LX, Wei YL, et al. Development of fluorescent aptasensing system for ultrasensitive analysis of kanamycin[J]. J Lumin, 2020, 222:117124.
doi: 10.1016/j.jlumin.2020.117124 URL |
| [42] |
Grate D, Wilson C. Laser-mediated, site-specific inactivation of RNA transcripts[J]. PNAS, 1999, 96(11):6131-6136.
pmid: 10339553 |
| [43] |
Rogers TA, Andrews GE, Jaeger L, et al. Fluorescent monitoring of RNA assembly and processing using the split-spinach aptamer[J]. ACS Synth Biol, 2015, 4(2):162-166.
doi: 10.1021/sb5000725 URL |
| [44] |
Song W, Filonov GS, Kim H, et al. Imaging RNA polymerase III transcription using a photostable RNA-fluorophore complex[J]. Nat Chem Biol, 2017, 13(11):1187-1194.
doi: 10.1038/nchembio.2477 URL |
| [45] |
Svensen N, Jaffrey SR. Fluorescent RNA aptamers as a tool to study RNA-modifying enzymes[J]. Cell Chem Biol, 2016, 23(3):415-425.
doi: 10.1016/j.chembiol.2015.11.018 pmid: 26877022 |
| [46] |
Paige JS, Nguyen-Duc T, Song W, et al. Fluorescence imaging of cellular metabolites with RNA[J]. Science, 2012, 335(6073):1194.
doi: 10.1126/science.1218298 URL |
| [47] |
Kim H, Jaffrey SR. A fluorogenic RNA-based sensor activated by metabolite-induced RNA dimerization[J]. Cell Chem Biol, 2019, 26(12):1725-1731.e6.
doi: 10.1016/j.chembiol.2019.09.013 URL |
| [48] |
Kellenberger CA, Wilson SC, Sales-Lee J, et al. RNA-based fluorescent biosensors for live cell imaging of second messengers cyclic di-GMP and cyclic AMP-GMP[J]. J Am Chem Soc, 2013, 135(13):4906-4909.
doi: 10.1021/ja311960g pmid: 23488798 |
| [49] | You M, Litke JL, Jaffrey SR. Imaging metabolite dynamics in living cells using a Spinach-based riboswitch[J]. PNAS, 2015, 112(21):E2756-E2765. |
| [50] |
Xu JS, Cotruvo JA. The czcD(NiCo)riboswitch responds to iron(II)[J]. Biochemistry, 2020, 59(15):1508-1516.
doi: 10.1021/acs.biochem.0c00074 URL |
| [51] |
Filonov GS, Kam CW, Song W, et al. In-gel imaging of RNA processing using broccoli reveals optimal aptamer expression strategies[J]. Chem Biol, 2015, 22(5):649-660.
doi: 10.1016/j.chembiol.2015.04.018 URL |
| [52] |
Ponchon L, Dardel F. Recombinant RNA technology:the tRNA scaffold[J]. Nat Methods, 2007, 4(7):571-576.
pmid: 17558412 |
| [53] |
Jung JK, Alam KK, Verosloff MS, et al. Cell-free biosensors for rapid detection of water contaminants[J]. Nat Biotechnol, 2020, 38(12):1451-1459.
doi: 10.1038/s41587-020-0571-7 URL |
| [54] |
Roxo C, Kotkowiak W, Pasternak A. G-quadruplex-forming aptamers—characteristics, applications, and perspectives[J]. Molecules, 2019, 24(20):3781.
doi: 10.3390/molecules24203781 URL |
| [55] |
Xiao XN, Zhu LJ, He WC, et al. Functional nucleic acids tailoring and its application[J]. Trac Trends Anal Chem, 2019, 118:138-157.
doi: 10.1016/j.trac.2019.05.027 URL |
| [56] |
Xu WT, He WC, Du ZH, et al. Functional nucleic acid nanomaterials:development, properties, and applications[J]. Angew Chem Int Ed, 2021, 60(13):6890-6918.
doi: 10.1002/anie.201909927 URL |
| [57] |
Sunbul M, Jäschke A. SRB-2:a promiscuous rainbow aptamer for live-cell RNA imaging[J]. Nucleic Acids Res, 2018, 46(18):e110.
doi: 10.1093/nar/gky543 URL |
| [58] |
Song W, Strack RL, Jaffrey SR. Imaging bacterial protein expression using genetically encoded RNA sensors[J]. Nat Methods, 2013, 10(9):873-875.
doi: 10.1038/nmeth.2568 URL |
| [1] | 李天顺, 李宸葳, 王佳, 朱龙佼, 许文涛. 功能核酸筛选过程中次级文库的有效制备[J]. 生物技术通报, 2023, 39(3): 116-122. |
| [2] | 刘宁宁, 王鑫昕, 兰欣悦, 褚华硕, 陈旭, 常世敏, 李腾飞, 许文涛. G-三链体可视化核酸传感器用于四环素的检测[J]. 生物技术通报, 2022, 38(10): 106-114. |
| [3] | 杨敏, 李舒婷, 杨文平, 李相阳, 许文涛. DNA/银纳米簇介导的功能核酸生物传感器研究进展[J]. 生物技术通报, 2020, 36(6): 245-254. |
| [4] | 肖冰, 罗云波, 黄昆仑, 张园, 许文涛. 功能核酸荧光标记型定量统一化检测技术的研究进展[J]. 生物技术通报, 2019, 35(7): 213-221. |
| [5] | 谢银侠, 王蔚然, 程楠, 许文涛. 电信号分子在电化学功能核酸生物传感器中的研究进展[J]. 生物技术通报, 2019, 35(5): 157-169. |
| [6] | 苏秋菊, 周翔, 李光鹏, 白春玲, 许文涛, 刘榜. MSTN基因编辑牛的鉴定及基因分型新方法[J]. 生物技术通报, 2019, 35(4): 208-212. |
| [7] | 肖冰, 刘榜, 罗云波, 黄昆仑, 张园, 李夏莹, 张秀杰, 许文涛, 周翔. 功能核酸荧光免标记型定量统一化检测技术的研究进展[J]. 生物技术通报, 2019, 35(3): 194-202. |
| [8] | 李宸葳, 杜再慧, 林少华, 罗云波, 许文涛. Pb2+功能核酸生物传感器的研究进展[J]. 生物技术通报, 2019, 35(1): 131-139. |
| [9] | 李凯, 罗云波, 许文涛. 10-23脱氧核酶介导的生物传感器研究进展[J]. 生物技术通报, 2019, 35(1): 140-150. |
| [10] | 刘星雨, 李春晖, 田晶晶, 邵向丽, 罗云波, 许文涛. 荧光铜纳米簇介导的生物传感器的研究进展[J]. 生物技术通报, 2019, 35(1): 170-186. |
| [11] | 张倩, 田晶晶, 罗云波, 许文涛. 功能核酸纳米机器生物传感器的研究进展[J]. 生物技术通报, 2018, 34(9): 2-14. |
| [12] | 张园, 肖冰, 田晶晶, 许文涛. 恒温技术介导的功能核酸生物传感器研究进展[J]. 生物技术通报, 2018, 34(9): 29-38. |
| [13] | 贺万崇, 黄昆仑, 许文涛. APE1介导的功能核酸生物传感器研究进展[J]. 生物技术通报, 2018, 34(9): 48-54. |
| [14] | 宋欢, 罗云波, 许文涛. 外切酶III介导的功能核酸生物传感器研究进展[J]. 生物技术通报, 2018, 34(9): 55-69. |
| [15] | 王联臻, 张倩, 李凯, 贺万崇, 罗云波, 黄昆仑, 许文涛. Mg2+功能核酸生物传感器检测技术的研究进展[J]. 生物技术通报, 2018, 34(9): 104-115. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||