








生物技术通报 ›› 2022, Vol. 38 ›› Issue (11): 151-161.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0251
收稿日期:2022-02-28
出版日期:2022-11-26
发布日期:2022-12-01
作者简介:党瑗,女,硕士研究生,研究方向:植物分子生物学;E-mail:基金资助:
DANG Yuan( ), LI Wei, MIAO Xiang, XIU Yu, LIN Shan-zhi(
), LI Wei, MIAO Xiang, XIU Yu, LIN Shan-zhi( )
)
Received:2022-02-28
Published:2022-11-26
Online:2022-12-01
摘要:
油体蛋白OLE在植物油脂合成累积中具有重要调控作用。依据山杏(Prunus sibirica)不同发育阶段种子的mRNA转录组测序数据,注释获得5个具有完整开放阅读框和典型保守结构域oleosin的油体蛋白OLE家族基因,通过差异转录谱分析及RT-qPCR检测,确定与山杏种子发育及油脂累积密切相关的高表达PsOLE4基因为研究对象,克隆该基因并进行生物信息学分析、组织特异表达检测、亚细胞定位分析、遗传转化拟南芥及其种子油脂含量和脂肪酸组分测定等研究。结果显示,PsOLE4基因全长序列为378 bp,可编码含125个氨基酸且分子量为13 kD的蛋白。该蛋白为疏水性无信号肽的非分泌蛋白,含有18个磷酸化位点、2个跨膜结构域和1个高度保守的脯氨酸结(PX5SP3P)结构域,定位于细胞膜上。PsOLE4基因在山杏种子中的表达量显著高于茎、叶及果实,而且PsOLE4基因表达可有效促进转基因拟南芥种子的脂肪酸含量提高与油脂累积,表明山杏PsOLE4基因具有种子表达特异性,对油脂累积具有重要调控作用。研究结果为后续开展山杏种子PsOLE4功能鉴定及应用奠定基础。
党瑗, 李维, 苗向, 修宇, 林善枝. 山杏油体蛋白基因PsOLE4克隆及其调控油脂累积功能分析[J]. 生物技术通报, 2022, 38(11): 151-161.
DANG Yuan, LI Wei, MIAO Xiang, XIU Yu, LIN Shan-zhi. Cloning of Oleosin Gene PsOLE4 from Prunus sibirica and Its Regulatory Function Analysis for Oil Accumulation[J]. Biotechnology Bulletin, 2022, 38(11): 151-161.
| 引物 Primer | 序列 Sequence(5'-3') | 备注 Annotation |
|---|---|---|
| PsOLE1-qf | CTTTGACCATCGCCACTCCG | PsOLE1基因荧光定量引物 |
| PsOLE1-qr | CGCCACACCGAATCCACC | |
| PsOLE2-qf | GAGACGGCGGGCTATTT | PsOLE2基因荧光定量引物 |
| PsOLE2-qr | CAGCATCCTGGGACTTATCTAC | |
| PsOLE3-qf | GCCTTCCCAGCCAACTTAT | PsOLE3基因荧光定量引物 |
| PsOLE3-qr | TTCTTGACTTCCGGTGGATTC | |
| PsOLE4-qf | ACGTTAACTGGGACAGTGATG | PsOLE4基因荧光定量引物 |
| PsOLE4-qr | GCTGTCAGGAACACGACTATAC | |
| PsOLE5-qf | TCTTGGGTCACCAACTACCT | PsOLE5基因荧光定量引物 |
| PsOLE5-qr | GTCCCACAAACTCCACCATATC | |
| CYP-qf | CAACGGATCTCAGTTCTTCGTCTGC | CYP内参基因荧光定量引物 |
| CYP-qr | GACCCAACCTTCTCGATGTTCTTCA | |
| UBC-qf | GAGACCAGCAATAACCGTGAA | UBC内参基因荧光定量引物 |
| UBC-qr | TCTTGTACTCCGTGGCATCCT | |
| PsOLE4-f1 | CTCTTTGAGCAACTAATGACGTACA | PsOLE4克隆引物 |
| PsOLE4-r1 | ATTATCCAAACAACCCAACTTACCC | |
| PsOLE4-f2 | TGGAGAGAACACGGGGGACTCTAGAATGGCTGATCAATCAAGACACGTC | PsOLE4亚细胞定位引物 |
| PsOLE4-r2 | GGTGGCGACCGGTACCCGGGGATCAAAAGGGCAGAAGTACTGCCCATAC | |
| PsOLE4-f3 | GAGAGAACACGGGGGACTCTAGAGATGGCTGATCAATCAAGACACGTC | PsOLE4表达载体引物 |
| PsOLE4-r3 | GAACGATCGGGGAAATTCGAGCTTCAAAAAGGGCAGAAGTACTGCCCA |
表1 实验所用引物及序列
Table 1 Primers and sequences used in the study
| 引物 Primer | 序列 Sequence(5'-3') | 备注 Annotation |
|---|---|---|
| PsOLE1-qf | CTTTGACCATCGCCACTCCG | PsOLE1基因荧光定量引物 |
| PsOLE1-qr | CGCCACACCGAATCCACC | |
| PsOLE2-qf | GAGACGGCGGGCTATTT | PsOLE2基因荧光定量引物 |
| PsOLE2-qr | CAGCATCCTGGGACTTATCTAC | |
| PsOLE3-qf | GCCTTCCCAGCCAACTTAT | PsOLE3基因荧光定量引物 |
| PsOLE3-qr | TTCTTGACTTCCGGTGGATTC | |
| PsOLE4-qf | ACGTTAACTGGGACAGTGATG | PsOLE4基因荧光定量引物 |
| PsOLE4-qr | GCTGTCAGGAACACGACTATAC | |
| PsOLE5-qf | TCTTGGGTCACCAACTACCT | PsOLE5基因荧光定量引物 |
| PsOLE5-qr | GTCCCACAAACTCCACCATATC | |
| CYP-qf | CAACGGATCTCAGTTCTTCGTCTGC | CYP内参基因荧光定量引物 |
| CYP-qr | GACCCAACCTTCTCGATGTTCTTCA | |
| UBC-qf | GAGACCAGCAATAACCGTGAA | UBC内参基因荧光定量引物 |
| UBC-qr | TCTTGTACTCCGTGGCATCCT | |
| PsOLE4-f1 | CTCTTTGAGCAACTAATGACGTACA | PsOLE4克隆引物 |
| PsOLE4-r1 | ATTATCCAAACAACCCAACTTACCC | |
| PsOLE4-f2 | TGGAGAGAACACGGGGGACTCTAGAATGGCTGATCAATCAAGACACGTC | PsOLE4亚细胞定位引物 |
| PsOLE4-r2 | GGTGGCGACCGGTACCCGGGGATCAAAAGGGCAGAAGTACTGCCCATAC | |
| PsOLE4-f3 | GAGAGAACACGGGGGACTCTAGAGATGGCTGATCAATCAAGACACGTC | PsOLE4表达载体引物 |
| PsOLE4-r3 | GAACGATCGGGGAAATTCGAGCTTCAAAAAGGGCAGAAGTACTGCCCA |

图1 山杏种子PsOLE蛋白保守结构域及PsOLE基因的动态转录表达水平分析 A:山杏种子PsOLE蛋白保守结构域分析;B:mRNA高通量测序分析PsOLE家族基因表达水平,以log10(FPKM)进行标准化;C:RT-qPCR测定PsOLE家族基因相对表达水平,以log10(相对表达量)进行标准化。图中误差线为标准差
Fig. 1 Analysis of conserved domain for PsOLEs and dynamic transcriptional expression of PsOLEs in P. sibirica seeds A:Analysis of conserved domain for PsOLE proteins in P. sibirica seeds. B:Expressions level of PsOLE family genes by mRNA high-throughput sequencing,the values were normalized as log10(FPKM). C:Determination of relative expressions of PsOLE family genes by RT-qPCR,the values were normalized as log10(relative expression). The bar in the figure refers to the standard deviation
图2 山杏种子PsOLE4基因的扩增电泳图谱 M:DNA marker 2000;1:PsOLE4的基因片段
Fig. 2 Amplification electrophoretic map of the PsOLE4 gene from P. sibirica seeds M:2000 DNA marker. 1:PsOLE4 gene fragment

图3 山杏种子PsOLE4蛋白生物信息学分析 A:山杏种子PsOLE4蛋白磷酸化位点分析;B:PsOLE4蛋白亲/疏水性分析;C:PsOLE4蛋白信号肽分析;D:PsOLE4蛋白跨膜结构分析;E:PsOLE4蛋白二级结构预测;F:PsOLE4蛋白三级结构预测
Fig. 3 Bioinformatics analysis for PSOLE4 protein from P. sibirica seeds A:Analysis of phosphorylation site for PsOLE4 protein from P. sibirica seeds. B:Analysis of hydrophilicity/hydrophobicity for PsOLE4. C:Analysis of signal peptide for PsOLE4. D:Analysis of transmembrane region for PsOLE4. E:Prediction of secondary structure for PsOLE4. F:Prediction of tertiary structure for PsOLE4
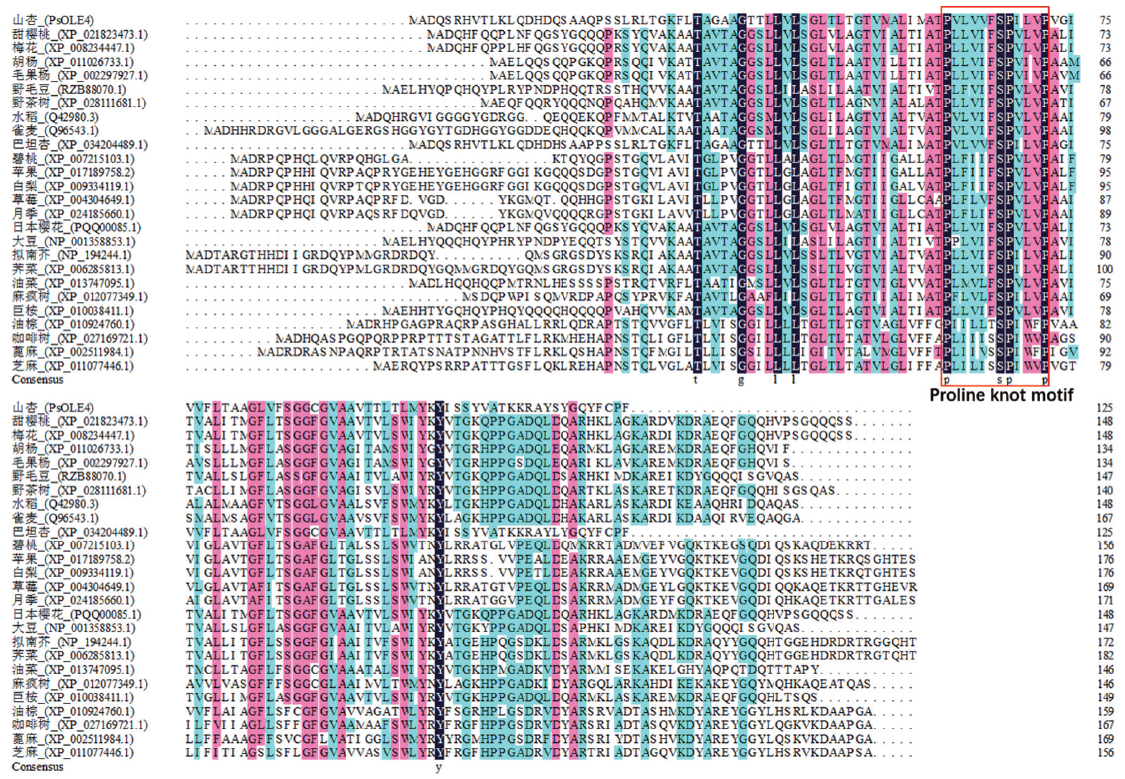
图4 山杏种子PsOLE4蛋白的多序列比对 山杏(Prunus sibirica),甜樱桃(Prunus mume),梅花(Prunus mume),胡杨(Populus euphratica),毛果杨(Populus trichocarpa),野毛豆(Glycine soja),野茶树(Camellia sinensis),水稻(Oryza sativa),雀麦(Bromus secalinus),巴坦杏(Prunus dulcis),碧桃(Prunus persica),苹果(Malus domestica),白梨(Pyrus × bretschneideri),草莓(Fragaria vesca),月季(Rosa chinensis),日本樱花(Prunus yedoensis),大豆(Glycine max),拟南芥(Arabidopsis thaliana),荠菜(Capsella rubella),油菜(Brassica napus),麻疯树(Jatropha curcas),巨桉(Eucalyptus grandis),油棕(Elaeis guineensis),咖啡树(Coffea eugenioides),蓖麻(Ricinus communis),芝麻(Sesamum indicum)
Fig. 4 Multiple sequence alignment of PsOLE4 protein from P. sibirica seeds
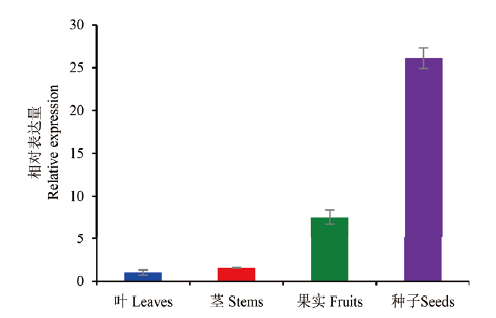
图6 山杏PsOLE4基因在不同组织中的相对表达量分析 以叶中PsOLE4基因的最低表达量为对照,标准化校正为1
Fig. 6 Analysis of relative expression for PsOLE4 gene in the different tissues of P. sibirica The lowest expression of PsOLE4 gene in the leaves was taken as control,and standardized calibration is 1

图7 山杏种子PsOLE4蛋白的亚细胞定位 Dark为荧光照片;Bright为明场照片;Merged为合并照片
Fig. 7 Subcellular localization of PsOLE4 from P. sibirica seeds Dark is defined as dark-field fluorescence image. Bright is defined as bright-field image. Merged is defined as the merged image in both bright and dark fields
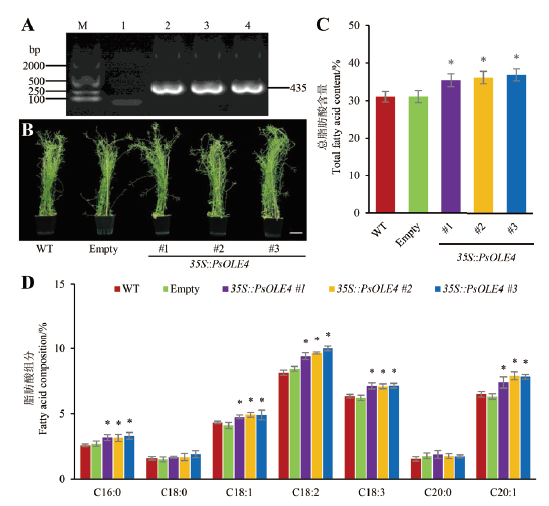
图8 转PsOLE4基因拟南芥的生长状态及其种子脂肪酸含量与组分分析 A:转基因拟南芥PCR鉴定,M:DNA marker,1:空载体植株(empty)PCR产物,2-4:转PsOLE4基因植株(35S∷PsOLE4 #1、35S∷PsOLE4 #2和35S∷PsOLE4 #3)PCR产物;B:野生型(WT)、空载体和过表达PsOLE4基因拟南芥生长状况,比例尺=5 cm;C:野生型、空载体和过表达PsOLE4基因拟南芥种子脂肪酸含量测定;D:拟南芥种子脂肪酸组分分析。星号(*)表示与野生型拟南芥差异显著P < 0.05。C16:0:棕榈酸;C18:0:硬脂酸;C18:1:油酸;C18:2:亚油酸;C18:3:亚麻酸;C20:0:花生酸;C20:1:花生油酸
Fig. 8 Analyses for growth status in PsOLE- transgenic A. thaliana and fatty acid content and composition of their seeds A:PCR identification for transgenic A. thaliana,M:DNA marker,1:PCR product from empty vector transgenic plant(empty),2-4:PCR product from PsOLE4 transgenic plants(35S∷PsOLE4 #1、35S∷PsOLE4 #2和35S∷PsOLE4 #3). B:Growth status of WT,empty and PsOLE4 overexpressing A. thaliana lines,scale bar=5 cm. C:Fatty acid contents in the seeds of WT,empty and PsOLE4 overexpressing transgenic A. thaliana lines. D:Fatty acid composition in seeds of A. thaliana. The asterisks(*)above the columns indicate significant differences at P<0.05. C16∶0:Palmitic acid. C18:0:Stearic acid. C18:1:Oleic acid. C18:2:Linoleic acid. C18:3:Linolenic acid. C20:0:Arachidic acid. C20:1:Eicosenoic acid
| [1] | 王利兵. 我国3种杏的地理分布及其植物学性状[J]. 林业科学研究, 2010, 23(3):435-439. |
| Wang LB. Geographic distribution and botanical characters of 3 Armeniaca plant in China[J]. For Res, 2010, 23(3):435-439. | |
| [2] |
刘潇菡, 林子欣, 修宇, 等. 山杏种子MATE家族分析及其重要成员MATE40克隆表达[J]. 生物技术通报, 2021, 37(11):197-211.
doi: 10.13560/j.cnki.biotech.bull.1985.2021-0590 |
| Liu XH, Lin ZX, Xiu Y, et al. Analysis of the MATE family in the seeds of Prunus sibirica and cloning and expression of its important member MATE40[J]. Biotechnol Bull, 2021, 37(11):197-211. | |
| [3] |
Guo JY, Li HP, Fan SQ, et al. Genetic variability of biodiesel properties in some Prunus L. (Rosaceae)species collected from Inner Mongolia, China[J]. Ind Crops Prod, 2015, 76:244-248.
doi: 10.1016/j.indcrop.2015.05.052 URL |
| [4] |
Wang LB, Yu HY. Biodiesel from Siberian apricot(Prunus sibirica L.)seed kernel oil[J]. Bioresour Technol, 2012, 112:355-358.
doi: 10.1016/j.biortech.2012.02.120 URL |
| [5] |
Niu J, An JY, Wang LB, et al. Transcriptomic analysis revealed the mechanism of oil dynamic accumulation during developing Siberian apricot(Prunus sibirica L.)seed kernels for the development of woody biodiesel[J]. Biotechnol Biofuels, 2015, 8:29.
doi: 10.1186/s13068-015-0213-3 URL |
| [6] |
Fan SQ, Liang TY, Yu HY, et al. Kernel characteristics, oil contents, fatty acid compositions and biodiesel properties in developing Siberian apricot(Prunus sibirica L.)seeds[J]. Ind Crops Prod, 2016, 89:195-199.
doi: 10.1016/j.indcrop.2016.05.012 URL |
| [7] | Niu J, Wang J, An JY, et al. Integrated mRNA and miRNA transcriptome reveal a cross-talk between developing response and hormone signaling for the seed kernels of Siberian apricot[J]. Sci Reports, 2016, 6:35675. |
| [8] |
Wang J, Lin WJ, Yin ZD, et al. Comprehensive evaluation of fuel properties and complex regulation of intracellular transporters for high oil production in developing seeds of Prunus sibirica for woody biodiesel[J]. Biotechnol Biofuels, 2019, 12:6.
doi: 10.1186/s13068-018-1347-x pmid: 30622648 |
| [9] |
Lin ZX, An JY, Wang J, et al. Integrated analysis of 454 and Illumina transcriptomic sequencing characterizes carbon flux and energy source for fatty acid synthesis in developing Lindera glauca fruits for woody biodiesel[J]. Biotechnol Biofuels, 2017, 10:134.
doi: 10.1186/s13068-017-0820-2 URL |
| [10] |
Bates PD, Stymne S, Ohlrogge J. Biochemical pathways in seed oil synthesis[J]. Curr Opin Plant Biol, 2013, 16(3):358-364.
doi: 10.1016/j.pbi.2013.02.015 pmid: 23529069 |
| [11] | Fang Y, Zhu RL, Mishler BD. Evolution of oleosin in land plants[J]. PLoS One, 2014, 9(8):e103806. |
| [12] |
Hyun TK, Kumar D, Cho YY, et al. Computational identification and phylogenetic analysis of the oil-body structural proteins, oleosin and caleosin, in Castor bean and flax[J]. Gene, 2013, 515(2):454-460.
doi: 10.1016/j.gene.2012.11.065 pmid: 23232356 |
| [13] |
Keddie JS, Hübner G, Slocombe SP, et al. Cloning and characterisation of an oleosin gene from Brassica napus[J]. Plant Mol Biol, 1992, 19(3):443-453.
pmid: 1377966 |
| [14] |
Chen JC, Lin RH, Huang HC, et al. Cloning, expression and isoform classification of a minor oleosin in sesame oil bodies[J]. J Biochem, 1997, 122(4):819-824.
pmid: 9399587 |
| [15] |
Vance VB, Huang AH. The major protein from lipid bodies of maize. Characterization and structure based on cDNA cloning[J]. J Biol Chem, 1987, 262(23):11275-11279.
pmid: 2440887 |
| [16] |
Huang NL, Huang MD, Chen TLL, et al. Oleosin of subcellular lipid droplets evolved in green algae[J]. Plant Physiol, 2013, 161(4):1862-1874.
doi: 10.1104/pp.112.212514 URL |
| [17] |
Kim HU, Hsieh K, Ratnayake C, et al. A novel group of oleosins is present inside the pollen of Arabidopsis[J]. J Biol Chem, 2002, 277(25):22677-22684.
doi: 10.1074/jbc.M109298200 URL |
| [18] | Zhang D, Zhang HY, Hu ZB, et al. Artificial selection on GmOLEO1 contributes to the increase in seed oil during soybean domestication[J]. PLoS Genet, 2019, 15(7):e1008267. |
| [19] |
Huang AHC. Plant lipid droplets and their associated proteins:potential for rapid advances[J]. Plant Physiol, 2018, 176(3):1894-1918.
doi: 10.1104/pp.17.01677 URL |
| [20] |
Huang MD, Huang AHC. Bioinformatics reveal five lineages of oleosins and the mechanism of lineage evolution related to structure/function from green algae to seed plants[J]. Plant Physiol, 2015, 169(1):453-470.
doi: 10.1104/pp.15.00634 URL |
| [21] | Beisson F, Ferté N, Bruley S, et al. Oil-bodies as substrates for lipolytic enzymes[J]. Biochim Biophys Acta, 2001, 1531(1/2):47-58. |
| [22] |
Miquel M, Trigui G, d’Andréa S, et al. Specialization of oleosins in oil body dynamics during seed development in Arabidopsis seeds[J]. Plant Physiol, 2014, 164(4):1866-1878.
doi: 10.1104/pp.113.233262 URL |
| [23] |
Shimada TL, Shimada T, Takahashi H, et al. A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana[J]. Plant J, 2008, 55(5):798-809.
doi: 10.1111/j.1365-313X.2008.03553.x URL |
| [24] |
Hsieh K, Huang AHC. Endoplasmic reticulum, oleosins, and oils in seeds and tapetum cells[J]. Plant Physiol, 2004, 136(3):3427-3434.
pmid: 15542496 |
| [25] |
Chen K, Yin YT, Liu S, et al. Genome-wide identification and functional analysis of oleosin genes in Brassica napus L.[J]. BMC Plant Biol, 2019, 19(1):294.
doi: 10.1186/s12870-019-1891-y URL |
| [26] | Niu J, Zhu BQ, Cai J, et al. Selection of reference genes for gene expression studies in Siberian Apricot(Prunus sibirica L.)Germplasm using quantitative real-time PCR[J]. PLoS One, 2014, 9(8):e103900. |
| [27] |
Clough SJ, Bent AF. Floral dip:a simplified method for Agrobac-terium-mediated transformation of Arabidopsis thaliana[J]. Plant J, 1998, 16(6):735-743.
pmid: 10069079 |
| [28] |
Zhang W, Wang SY, Yu FW, et al. Genome-wide identification and expression profiling of sugar transporter protein(STP)family genes in cabbage(Brassica oleracea var. capitata L.)reveals their involvement in clubroot disease responses[J]. Genes, 2019, 10(1):71.
doi: 10.3390/genes10010071 URL |
| [29] | Li LK, Wang ZR, Li YP, et al. Characterization of genes encoding ω-6 desaturase PoFAD2 and PoFAD6, and ω-3 desaturase PoFAD3 for ALA accumulation in developing seeds of oil crop Paeonia ostii var. lishizhenii[J]. Plant Sci, 2021, 312:111029. |
| [30] |
Ojha R, Kaur S, Sinha K, et al. Characterization of oleosin genes from forage sorghum in Arabidopsis and yeast reveals their role in storage lipid stability[J]. Planta, 2021, 254(5):97.
doi: 10.1007/s00425-021-03744-8 URL |
| [31] |
Song YH, Wang XD, Rose RJ. Oil body biogenesis and biotechnology in legume seeds[J]. Plant Cell Rep, 2017, 36(10):1519-1532.
doi: 10.1007/s00299-017-2201-5 pmid: 28866824 |
| [32] | 徐赫, 潘丽娟, 陈明娜, 等. 花生油质蛋白基因的克隆与表达分析[J]. 花生学报, 2019, 48(3):9-14. |
| Xu H, Pan LJ, Chen MN, et al. Cloning and expression analysis of oleosin genes in peanut[J]. J Peanut Sci, 2019, 48(3):9-14. | |
| [33] | 宋健, 熊宏, 余进德, 等. 麻疯树油质蛋白JcOle14.3基因克隆及序列分析[J]. 中南林业科技大学学报, 2016, 36(6):15-22. |
| Song J, Xiong H, Yu JD, et al. Cloning and sequence analysis of oleosin gene JcOle14.3 in Jatropha curcas[J]. J Central South Univ For Technol, 2016, 36(6):15-22. | |
| [34] |
Abenes M, Holbrook L, Moloney M. Transient expression and oil body targeting of an Arabidopsis oleosin-GUS reporter fusion protein in a range of oilseed embryos[J]. Plant Cell Rep, 1997, 17(1):1-7.
doi: 10.1007/s002990050341 pmid: 30732411 |
| [35] |
Crowe AJ, Abenes M, Plant A, et al. The seed-specific transactivator, ABI3, induces oleosin gene expression[J]. Plant Sci, 2000, 151(2):171-181.
pmid: 10808073 |
| [36] |
Yang Z, Liu XL, Wang K, et al. ABA-INSENSITIVE 3 with or without FUSCA3 highly up-regulates lipid droplet proteins and activates oil accumulation[J]. J Exp Bot, 2021, 73(7):2077-2092.
doi: 10.1093/jxb/erab524 URL |
| [37] |
Chung KJ, Hwang SK, Hahn BS, et al. Authentic seed-specific activity of the Perilla oleosin 19 gene promoter in transgenic Arabidopsis[J]. Plant Cell Rep, 2008, 27(1):29-37.
doi: 10.1007/s00299-007-0440-6 URL |
| [38] |
Kohno-Murase J, Iwabuchi M, Endo-Kasahara S, et al. Production of trans-10, cis-12 conjugated linoleic acid in rice[J]. Transgenic Res, 2006, 15(1):95-100.
pmid: 16475013 |
| [39] |
Liu WX, Liu HL, Qu LQ. Embryo-specific expression of soybean oleosin altered oil body morphogenesis and increased lipid content in transgenic rice seeds[J]. Theor Appl Genet, 2013, 126(9):2289-2297.
doi: 10.1007/s00122-013-2135-4 pmid: 23748707 |
| [40] |
Lu YB, Chi MH, Li LX, et al. Genome-wide identification, expression profiling, and functional validation of Oleosin gene family in Carthamus tinctorius L[J]. Front Plant Sci, 2018, 9:1393.
doi: 10.3389/fpls.2018.01393 URL |
| [41] |
Yuan YC, Cao XZ, Zhang HJ, et al. Genome-wide identification and analysis of Oleosin gene family in four cotton species and its involvement in oil accumulation and germination[J]. BMC Plant Biol, 2021, 21(1):569.
doi: 10.1186/s12870-021-03358-y URL |
| [1] | 王贵芳, 姚元涛, 许海峰, 相昆, 梁家慧, 张淑辉, 王文茹, 张明娟, 张美勇, 陈新. 核桃JrSnRK1α1.1调控种子油脂合成与积累[J]. 生物技术通报, 2023, 39(9): 183-191. |
| [2] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [3] | 李博, 刘合霞, 陈宇玲, 周兴文, 朱宇林. 金花茶CnbHLH79转录因子的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 241-250. |
| [4] | 胡明月, 杨宇, 郭仰东, 张喜春. 低温胁迫下番茄SlMYB96的功能分析[J]. 生物技术通报, 2023, 39(4): 236-245. |
| [5] | 黄文莉, 李香香, 周炆婷, 罗莎, 姚维嘉, 马杰, 张芬, 沈钰森, 顾宏辉, 王建升, 孙勃. 利用CRISPR/Cas9技术靶向编辑青花菜BoZDS[J]. 生物技术通报, 2023, 39(2): 80-87. |
| [6] | 杨旭妍, 赵爽, 马天意, 白玉, 王玉书. 三个甘蓝WRKY基因的克隆及其对非生物胁迫的表达[J]. 生物技术通报, 2023, 39(11): 261-269. |
| [7] | 张玉娟, 黎冬华, 宫慧慧, 崔新晓, 高春华, 张秀荣, 游均, 赵军胜. 芝麻NAC转录因子基因SiNAC77的克隆及耐盐功能分析[J]. 生物技术通报, 2023, 39(11): 308-317. |
| [8] | 陈浩婷, 张玉静, 刘洁, 代泽敏, 刘伟, 石玉, 张毅, 李天来. 低磷胁迫下番茄转录因子WRKY6功能分析[J]. 生物技术通报, 2023, 39(10): 136-147. |
| [9] | 周家燕, 邹建, 陈卫英, 吴一超, 陈奚潼, 王倩, 曾文静, 胡楠, 杨军. 植物多基因干扰载体体系构建与效用分析[J]. 生物技术通报, 2023, 39(1): 115-126. |
| [10] | 孙威, 张艳, 王聿晗, 徐僡, 徐小蓉, 鞠志刚. 马缨杜鹃Rd3GT1的克隆及对矮牵牛花色形成的影响[J]. 生物技术通报, 2022, 38(9): 198-206. |
| [11] | 郭志浩, 金泽鑫, 刘琦, 高利. 小麦矮腥黑粉菌效应蛋白g11335的生物信息学分析、亚细胞定位及毒性验证[J]. 生物技术通报, 2022, 38(8): 110-117. |
| [12] | 于秋琳, 马婧怡, 赵盼, 孙鹏芳, 何玉美, 刘世彪, 郭惠红. 绞股蓝GpMIR156a和GpMIR166b的克隆与功能分析[J]. 生物技术通报, 2022, 38(7): 186-193. |
| [13] | 陆新华, 孙德权, 张秀梅. 介孔硅纳米粒作为植物细胞转基因载体的研究[J]. 生物技术通报, 2022, 38(7): 194-204. |
| [14] | 杨佳宝, 周至铭, 张展, 冯丽, 孙黎. 向日葵HaLACS1的克隆、表达及酵母功能互补鉴定[J]. 生物技术通报, 2022, 38(6): 147-156. |
| [15] | 镐青青, 姚圣, 刘佳禾, 陈佩珍, 张梦洋, 季孔庶. 马尾松NAC转录因子基因PmNAC8的克隆及表达分析[J]. 生物技术通报, 2022, 38(4): 202-216. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||