








生物技术通报 ›› 2023, Vol. 39 ›› Issue (8): 204-212.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0098
王佳蕊1,2( ), 孙培媛1,2, 柯瑾1,2, 冉彬1,2, 李洪有1(
), 孙培媛1,2, 柯瑾1,2, 冉彬1,2, 李洪有1( )
)
收稿日期:2023-02-08
出版日期:2023-08-26
发布日期:2023-09-05
通讯作者:
李洪有,男,博士,教授,研究方向:荞麦分子育种;E-mail: lihongyouluod@163.com作者简介:王佳蕊,女,硕士研究生,研究方向:荞麦分子生物学;E-mail: wangjiarui803@163.com
基金资助:
WANG Jia-rui1,2( ), SUN Pei-yuan1,2, KE Jin1,2, RAN Bin1,2, LI Hong-you1(
), SUN Pei-yuan1,2, KE Jin1,2, RAN Bin1,2, LI Hong-you1( )
)
Received:2023-02-08
Published:2023-08-26
Online:2023-09-05
摘要:
苦荞[Fagopyrum tataricum(L.)Gaertn]富含类黄酮C-糖苷,具有抗氧化、抗癌和消炎等多种保健作用。通过RT-PCR(reverse transcription-polymerase chain reaction)技术从苦荞中克隆得到了一个糖基转移酶基因,命名为FtUGT143,对其进行生物信息学、分子对接、基因表达、基因表达量与代谢物含量相关性等分析。结果表明,FtUGT143全长CDS序列为678 bp,编码226个氨基酸。密码子偏好分析结果显示,FtUGT143具有双子叶植物的密码子使用偏好性。多序列比对和进化树分析表明,FtUGT143是植物糖基转移酶基因家族中的C-糖基转移酶亚家族成员。分子对接结果表明,FtUGT143能与合成黄酮C-糖苷(牡荆素、异牡荆素、荭草素和异荭草素)的底物芹菜素和木犀草素相互作用。RT-qPCR结果显示,FtUGT143基因在苦荞的各个组织部位中均有表达,但在芽苗期的根、茎、叶中显著表达,且其在不同组织部位的表达量与4种黄酮C-糖苷积累量具有较好的相关性。研究结果表明FtUGT143是C-糖基转移酶,它可能参与苦荞中黄酮C-糖苷生物合成,对于揭示苦荞中黄酮C-糖苷的合成机制具有重要意义。
王佳蕊, 孙培媛, 柯瑾, 冉彬, 李洪有. 苦荞糖基转移酶基因FtUGT143的克隆及表达分析[J]. 生物技术通报, 2023, 39(8): 204-212.
WANG Jia-rui, SUN Pei-yuan, KE Jin, RAN Bin, LI Hong-you. Cloning and Expression Analyses of C-glycosyltransferase Gene FtUGT143 in Fagopyrum tataricum[J]. Biotechnology Bulletin, 2023, 39(8): 204-212.
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 用途Use |
|---|---|---|
| FtUGT143-F | ATGGACTGGATGGATGAGATGCTA | 基因克隆Gene cloning |
| FtUGT143-R | CTGTCAAATTTCGACACTGTCAAG | 基因克隆Gene cloning |
| qFtUGT143-F | TAAGGAGATGGCTGAAGCTG | 荧光定量Fluorescent quantification |
| qFtUGT143-R | CACCGAACTTGGATATCCGA | 荧光定量Fluorescent quantification |
| qFtUPL7-F | TTCACGGGCACCATTACTGG | 荧光定量内参Fluorescent quantification of internal reference primers |
| qFtUPL7-R | AGGTGGAAGCTGAAGGAAGC | 荧光定量内参Fluorescent quantification of internal reference primers |
表1 本研究中所用引物序列
Table 1 List of primers'sequences in this study
| 引物名称Primer name | 引物序列Primer sequence(5'-3') | 用途Use |
|---|---|---|
| FtUGT143-F | ATGGACTGGATGGATGAGATGCTA | 基因克隆Gene cloning |
| FtUGT143-R | CTGTCAAATTTCGACACTGTCAAG | 基因克隆Gene cloning |
| qFtUGT143-F | TAAGGAGATGGCTGAAGCTG | 荧光定量Fluorescent quantification |
| qFtUGT143-R | CACCGAACTTGGATATCCGA | 荧光定量Fluorescent quantification |
| qFtUPL7-F | TTCACGGGCACCATTACTGG | 荧光定量内参Fluorescent quantification of internal reference primers |
| qFtUPL7-R | AGGTGGAAGCTGAAGGAAGC | 荧光定量内参Fluorescent quantification of internal reference primers |
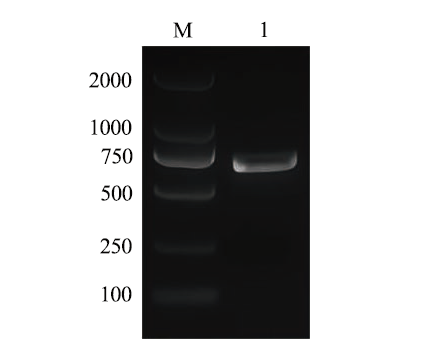
图1 苦荞FtUGT143基因全长CDS克隆 M:DNA maker 2000;1:FtUGT143的扩增片段
Fig. 1 Full-length CDS clone of the FtUGT143 gene of buckwheat(Fagopyrum tataricum) M: DNA maker 2000. 1: Amplified fragment of FtUGT143
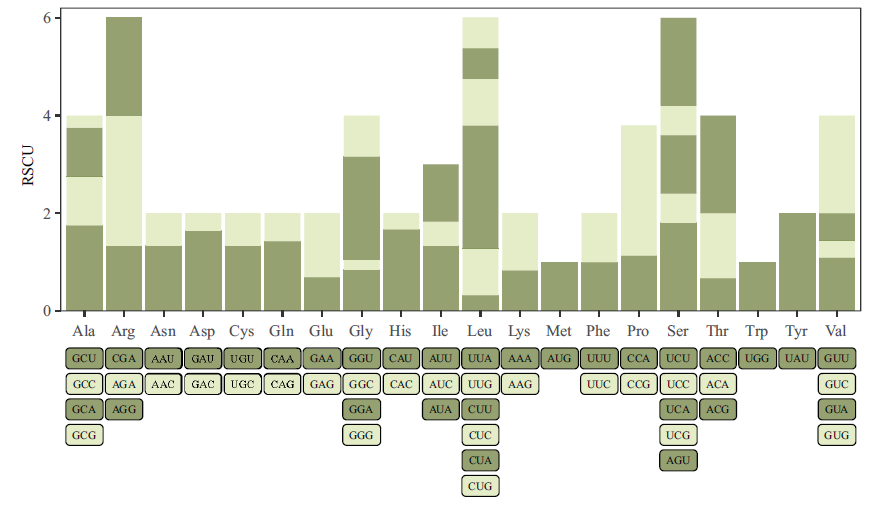
图2 FtUGT143密码子RSCU值分析 横坐标下方的图例分别对应上方该密码子的RSCU值(反映密码子实际出现次数与预期出现次数的比例关系)
Fig. 2 Analysis of FtUGT143 codon's RSCU values The legend below the abscissa axis corresponds to the RSCU value of the codon above(reflecting the proportional relationship between the actual occurrence times of the codon and the expected occurrence times)
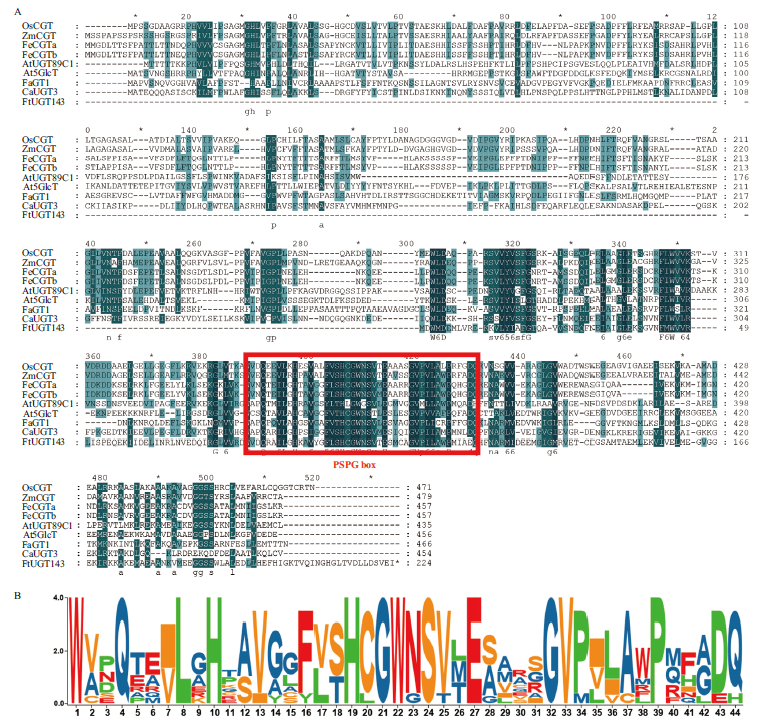
图3 植物UGT蛋白氨基酸的序列多重比对与PSPG box比对分析 A:植物UGT蛋白氨基酸的序列多重比对(FtUGT143:苦荞;OsCGT(XP_015641684.1):水稻;ZmCGT(PWZ07394.1):玉米;FeCGTa(A0A0A1HA03.1):甜荞;FeCGTb(A0A0A1H7N4.1):甜荞;GmUGT708D1(NP_001347241.1):大豆;At5GlcT(NP_193146.1):拟南芥;FaGT1(AAU09442.1):草莓;CaUGT3(BAH80312.1):长春花); 红框标注为糖基转移酶保守序列的PSPG box;B:PSPG box比对分析
Fig. 3 Sequence multi-alignment of plant UGT protein amino acids and comparative analysis of PSPG box A: Multiple Alignment of amino acid sequences of plant UGT protein(FtUGT143: Fagopyrum tataricum. OsCGT(XP_015641684.1): Oryza sativa; ZmCGT(PWZ07394.1): Zea mays; FeCGTa(A0A0A1HA03.1): Fagopyrum esculentum; FeCGTb(A0A0A1H7N4.1): Fagopyrum esculentum; GmUGT708D1(NP_001347241.1): Glycine max; At5GlcT(NP_193146.1): Arabidopsis thaliana; FaGT1(AAU09442.1): Fragaria × ananassa; CaUGT3(BAH80312.1): Catharanthus roseus). The red box indicates the PSPG box, a conserved sequence motif found in glycosyltransferases. B: Comparative analysis of PSPG box
| 配体小分子 Ligand small molecule | 结合能 Binding energy/(kJ·mol-1) |
|---|---|
| 山奈酚 Kaempferol | - 29.706 4 |
| 表儿茶素 Epicatechin | - 31.796 4 |
| 木犀草素 Luteolin | - 33.890 4 |
| 芹菜素 Apigenin | - 32.216 8 |
表2 FtUGT143与4种类黄酮代谢小分子的对接结果
Table 2 Docking results of FtUGT143 with four flavonoids metabolizing small molecules
| 配体小分子 Ligand small molecule | 结合能 Binding energy/(kJ·mol-1) |
|---|---|
| 山奈酚 Kaempferol | - 29.706 4 |
| 表儿茶素 Epicatechin | - 31.796 4 |
| 木犀草素 Luteolin | - 33.890 4 |
| 芹菜素 Apigenin | - 32.216 8 |
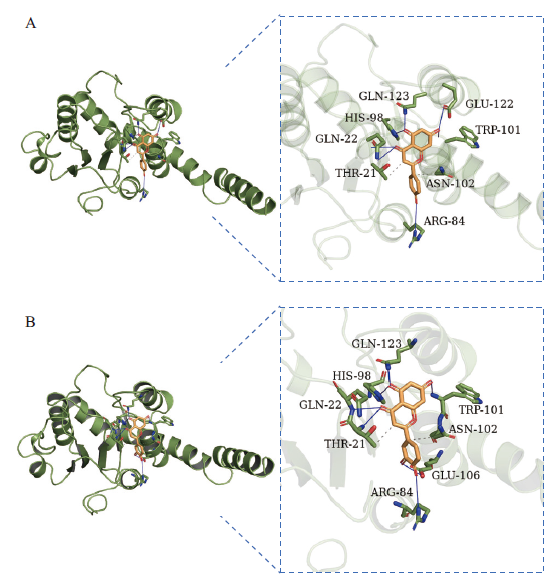
图5 FtUGT143和木犀草素、芹菜素对接示意图 A:FtUGT143和木犀草素对接示意图;B:FtUGT143和芹菜素对接示意图
Fig. 5 Schematic diagram of FtUGT143 docking with luteolin and apigenin A: Schematic diagram of FtUGT143 and luteolin docking. B: Schematic diagram of FtUGT143 and apigenin docking
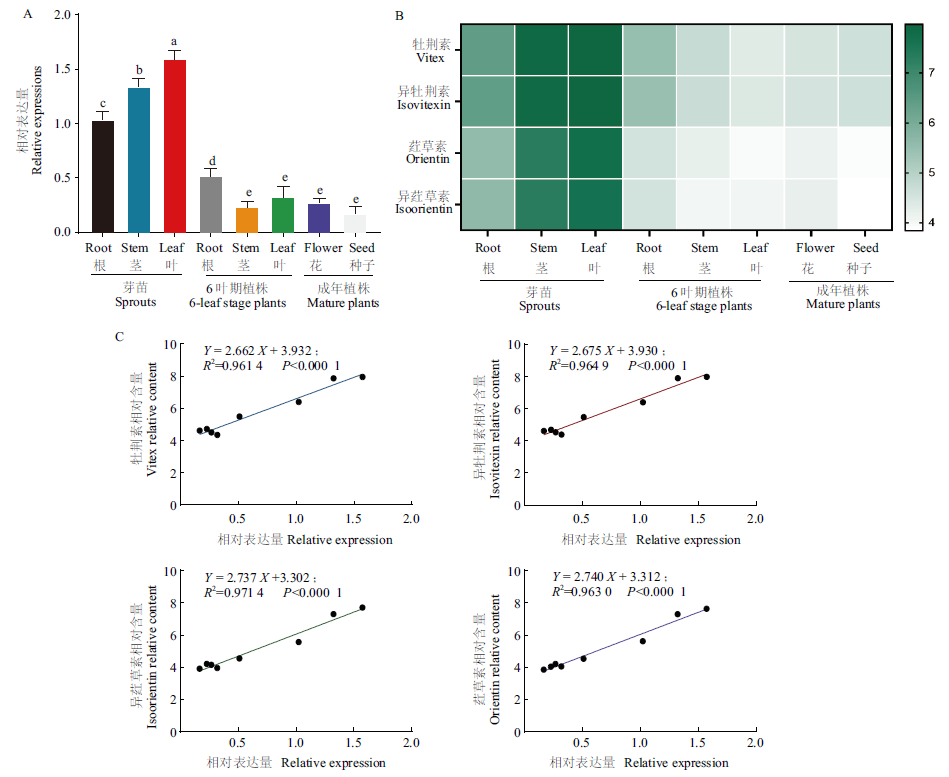
图6 FtUGT143在苦荞不同组织部位中的表达量与4种C-黄酮含量间的相关性 A:FtUGT143在不同组织部位中的表达;B:4种C-黄酮在不同组织部位的相对含量;C:FtUGT143在不同组织部位表达量与4种C-黄酮含量间的相关性
Fig. 6 Correlation between the expressions of FtUGT143 in the different tissue sites of buckwheat and the content of four C-flavonoids A: Expression of FtUGT143 in different tissue sites. B: Relative content of four C-flavonoids in different tissue sites. C: Correlation between the expression of FtUGT143 and the content of four C-flavonoids in different tissue sites
| [1] | Baskar V, Venkatesh R, Ramalingam S. Flavonoids(antioxidants systems)in higher plants and their response to stresses[M]// Antioxidants and Antioxidant Enzymes in Higher Plants. Cham: Springer, 2018: 253-268. |
| [2] | Kumar V, Suman U, et al. Flavonoid secondary metabolite: biosynthesis and role in growth and development in plants[M]// Recent Trends and Techniques in Plant Metabolic Engineering. Singapore: Springer, 2018: 19-45. |
| [3] |
Li HY, Lyu QY, Liu AK, et al. Comparative metabolomics study of Ta-rtary(Fagopyrum tataricum(L.)Gaertn)and common(Fagopyrum esculentum Moench)buckwheat seeds[J]. Food Chem, 2022, 371: 131125.
doi: 10.1016/j.foodchem.2021.131125 URL |
| [4] |
Huda MN, Lu S, Jahan T, et al. Treasure from garden: Bioactive compounds of buckwheat[J]. Food Chem, 2021, 335: 127653.
doi: 10.1016/j.foodchem.2020.127653 URL |
| [5] |
解林峰, 任传宏, 张波, 等. 植物类黄酮生物合成相关UDP-糖基转移酶研究进展[J]. 园艺学报, 2019, 46(9): 1655-1669.
doi: 10.16420/j.issn.0513-353x.2019-0237 |
|
Xie LF, Ren CH, Zhang B, et al. Plant UDP-glycosyltransferases in flavonoids biosynthesis[J]. Acta Hortic Sin, 2019, 46(9): 1655-1669.
doi: 10.16420/j.issn.0513-353x.2019-0237 |
|
| [6] |
Dai LH, Hu YM, Chen CC, et al. Flavonoid C-glycosyltransferases: function, evolutionary relationship, catalytic mechanism and protein engineering[J]. Chembioeng Rev, 2021, 8(1): 15-26.
doi: 10.1002/cben.v8.1 URL |
| [7] | Ren GL, Zhong Y, Ke G, et al. The mechanism of compound Anshen essential oil in the treatment of insomnia was examined by network pharmacology[J]. Evid Based Complement Alternat Med, 2019, 2019: 9241403. |
| [8] | He JB, Zhao P, Hu ZM, et al. Molecular and structural characterization of a promiscuous C-glycosyltransferase from Trollius chinensis[J]. Angewandte Chemie, 2019, 131(33): 11637-11644. |
| [9] |
Wang X, Li CF, Zhou C, et al. Molecular characterization of the C-glucosylation for puerarin biosynthesis in Pueraria lobata[J]. Plant J, 2017, 90(3): 535-546.
doi: 10.1111/tpj.13510 URL |
| [10] |
Mashima K, Hatano M, Suzuki H, et al. Identification and characterization of apigenin 6-C-glucosyltransferase involved in biosynthesis of isosaponarin in wasabi(Eutrema japonicum)[J]. Plant Cell Physiol, 2019, 60(12): 2733-2743.
doi: 10.1093/pcp/pcz164 pmid: 31418788 |
| [11] |
Brazier-Hicks M, Evans KM, Gershater MC, et al. The C-glycosylation of flavonoids in cereals[J]. J Biol Chem, 2009, 284(27): 17926-17934.
doi: 10.1074/jbc.M109.009258 pmid: 19411659 |
| [12] |
Nagatomo Y, Usui S, Ito T, et al. Purification, molecular cloning and functional characterization of flavonoid C-glucosyltransferases from Fagopyrum esculentum M.(buckwheat)cotyledon[J]. Plant J, 2014, 80(3): 437-448.
doi: 10.1111/tpj.2014.80.issue-3 URL |
| [13] |
Yao PF, Deng RY, Huang YJ, et al. Diverse biological effects of glycosyltransferase genes from Tartary buckwheat[J]. BMC Plant Biol, 2019, 19(1): 339.
doi: 10.1186/s12870-019-1955-z pmid: 31382883 |
| [14] |
Yin QG, Han XY, Han ZX, et al. Genome-wide analyses reveals a glucosyltransferase involved in rutin and emodin glucoside biosynthesis in Tartary buckwheat[J]. Food Chem, 2020, 318: 126478.
doi: 10.1016/j.foodchem.2020.126478 URL |
| [15] |
Zhou J, Li CL, Gao F, et al. Characterization of three glucosyltransferase genes in Tartary buckwheat and their expression after cold stress[J]. J Agric Food Chem, 2016, 64(37): 6930-6938.
doi: 10.1021/acs.jafc.6b02064 URL |
| [16] |
Zhang KX, He M, Fan Y, et al. Resequencing of global Tartary buckwheat accessions reveals multiple domestication events and key loci associated with agronomic traits[J]. Genome Biol, 2021, 22(1): 23.
doi: 10.1186/s13059-020-02217-7 pmid: 33430931 |
| [17] |
Xu HT, Jiang ZQ, Lin ZM, et al. FtUGT79A15 is responsible for rutinosylation in flavonoid diglycoside biosynthesis in Fagopyrum tataricum[J]. Plant Physiol Biochem, 2022, 181: 33-41.
doi: 10.1016/j.plaphy.2022.04.004 URL |
| [18] |
Li HY, Lyu QY, Ma C, et al. Metabolite profiling and transcriptome analyses provide insights into the flavonoid biosynthesis in the developing seed of Tartary buckwheat(Fagopyrum tataricum)[J]. J Agric Food Chem, 2019, 67(40): 11262-11276.
doi: 10.1021/acs.jafc.9b03135 URL |
| [19] | 王璐瑗, 荣玉萍, 黄娟, 等. 211份金荞麦收集系根茎黄酮含量的分析评价[J]. 贵州师范大学学报: 自然科学版, 2019, 37(4): 25-30, 48. |
| Wang LY, Rong YP, Huang J, et al. Analysis and evaluation of the flavonoid content of rhizomes of 211 different Golden buckwheat accessions(Fagopyrum cymosum complex)[J]. J Guizhou Norm Univ Nat Sci, 2019, 37(4): 25-30, 48. | |
| [20] |
姚宇, 顾佳珺, 孙超, 等. 植物类黄酮UDP-糖基转移酶研究进展[J]. 生物技术通报, 2022, 38(12): 47-57.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-0236 |
|
Yao Y, Gu JJ, Sun C, et al. Advances in plant flavonoids UDP-glycosyltransferase[J]. Biotechnol Bull, 2022, 38(12): 47-57.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-0236 |
|
| [21] |
Peng M, Shahzad R, Gul A, et al. Differentially evolved glucosyltransferases determine natural variation of rice flavone accumulation and UV-tolerance[J]. Nat Commun, 2017, 8(1): 1975.
doi: 10.1038/s41467-017-02168-x pmid: 29213047 |
| [22] |
Masada S, Terasaka K, Oguchi Y, et al. Functional and structural characterization of a flavonoid glucoside 1, 6-glucosyltransferase from Catharanthus roseus[J]. Plant Cell Physiol, 2009, 50(8): 1401-1415.
doi: 10.1093/pcp/pcp088 URL |
| [23] | 赵蕾. 毛竹中黄酮-C-糖基转移酶基因的克隆和功能鉴定[D]. 北京: 中国林业科学研究院, 2019. |
| Zhao L. Cloning and functional identification of flavonoid-C-glycosyltransferase genes in Phyllostachys edulis[D]. Beijing: Chinese Academy of Forestry, 2019. | |
| [24] | 付炎, 王于方, 李力更, 等. 天然药物化学史话: 天然产物研究与诺贝尔奖[J]. 中草药, 2016, 47(21): 3749-3765. |
| Fu Y, Wang YF, Li LG, et al. Historical story on natural medicinal chemistry: nature products research and Nobel Prize[J]. Chin Tradit Herb Drugs, 2016, 47(21): 3749-3765. | |
| [25] |
Vogt T, Jones P. Glycosyltransferases in plant natural product synthesis: characterization of a supergene family[J]. Trends Plant Sci, 2000, 5(9): 380-386.
doi: 10.1016/s1360-1385(00)01720-9 pmid: 10973093 |
| [1] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [2] | 孙明慧, 吴琼, 刘丹丹, 焦小雨, 王文杰. 茶树CsTMFs的克隆与表达分析[J]. 生物技术通报, 2023, 39(7): 151-159. |
| [3] | 赵雪婷, 高利燕, 王俊刚, 沈庆庆, 张树珍, 李富生. 甘蔗AP2/ERF转录因子基因ShERF3的克隆、表达及其编码蛋白的定位[J]. 生物技术通报, 2023, 39(6): 208-216. |
| [4] | 刘辉, 卢扬, 叶夕苗, 周帅, 李俊, 唐健波, 陈恩发. 外源硫诱导苦荞镉胁迫响应的比较转录组学分析[J]. 生物技术通报, 2023, 39(5): 177-191. |
| [5] | 姜晴春, 杜洁, 王嘉诚, 余知和, 王允, 柳忠玉. 虎杖转录因子PcMYB2的表达特性和功能分析[J]. 生物技术通报, 2023, 39(5): 217-223. |
| [6] | 姚姿婷, 曹雪颖, 肖雪, 李瑞芳, 韦小妹, 邹承武, 朱桂宁. 火龙果溃疡病菌实时荧光定量PCR内参基因的筛选[J]. 生物技术通报, 2023, 39(5): 92-102. |
| [7] | 王艺清, 王涛, 韦朝领, 戴浩民, 曹士先, 孙威江, 曾雯. 茶树SMAS基因家族的鉴定及互作分析[J]. 生物技术通报, 2023, 39(4): 246-258. |
| [8] | 刘思佳, 王浩楠, 付宇辰, 闫文欣, 胡增辉, 冷平生. ‘西伯利亚’百合LiCMK基因克隆及功能分析[J]. 生物技术通报, 2023, 39(3): 196-205. |
| [9] | 王涛, 漆思雨, 韦朝领, 王艺清, 戴浩民, 周喆, 曹士先, 曾雯, 孙威江. CsPPR和CsCPN60-like在茶树白化叶片中的表达分析及互作蛋白验证[J]. 生物技术通报, 2023, 39(3): 218-231. |
| [10] | 庞强强, 孙晓东, 周曼, 蔡兴来, 张文, 王亚强. 菜心BrHsfA3基因克隆及其对高温胁迫的响应[J]. 生物技术通报, 2023, 39(2): 107-115. |
| [11] | 苗淑楠, 高宇, 李昕儒, 蔡桂萍, 张飞, 薛金爱, 季春丽, 李润植. 大豆GmPDAT1参与油脂合成和非生物胁迫应答的功能分析[J]. 生物技术通报, 2023, 39(2): 96-106. |
| [12] | 葛雯冬, 王腾辉, 马天意, 范震宇, 王玉书. 结球甘蓝PRX基因家族全基因组鉴定与逆境条件下的表达分析[J]. 生物技术通报, 2023, 39(11): 252-260. |
| [13] | 杨旭妍, 赵爽, 马天意, 白玉, 王玉书. 三个甘蓝WRKY基因的克隆及其对非生物胁迫的表达[J]. 生物技术通报, 2023, 39(11): 261-269. |
| [14] | 陈楚怡, 杨小梅, 陈胜艳, 陈斌, 岳莉然. ABA和干旱胁迫下菊花脑ZF-HD基因家族的表达分析[J]. 生物技术通报, 2023, 39(11): 270-282. |
| [15] | 尤垂淮, 谢津津, 张婷, 崔天真, 孙欣路, 臧守建, 武奕凝, 孙梦瑶, 阙友雄, 苏亚春. 钩吻脂氧合酶基因 GeLOX1 的鉴定及低温胁迫表达分析[J]. 生物技术通报, 2023, 39(11): 318-327. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||