








生物技术通报 ›› 2023, Vol. 39 ›› Issue (8): 213-219.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0128
徐靖1( ), 朱红林1, 林延慧1, 唐力琼1, 唐清杰1,2, 王效宁1,2(
), 朱红林1, 林延慧1, 唐力琼1, 唐清杰1,2, 王效宁1,2( )
)
收稿日期:2023-02-15
出版日期:2023-08-26
发布日期:2023-09-05
通讯作者:
王效宁,男,硕士,研究员,研究方向:粮食作物种质资源创新利用;E-mail: wxning2599@163.com作者简介:徐靖,女,硕士,研究员,研究方向:甘薯种质资源创新利用;E-mail: xujing6732807@126.com
基金资助:
XU Jing1( ), ZHU Hong-lin1, LIN Yan-hui1, TANG Li-qiong1, TANG Qing-jie1,2, WANG Xiao-ning1,2(
), ZHU Hong-lin1, LIN Yan-hui1, TANG Li-qiong1, TANG Qing-jie1,2, WANG Xiao-ning1,2( )
)
Received:2023-02-15
Published:2023-08-26
Online:2023-09-05
摘要:
羟基肉桂酰辅酶A奎尼酸羟基肉桂酰转移酶(hydroxycinnamoyl CoA quinate hydroxycinnamoyl transferase, HQT)是绿原酸生物合成的最后一步限速酶。IbHQT1是甘薯绿原酸生物合成的关键HQT基因,为进一步揭示HbHQT1在甘薯绿原酸生物合成中的作用和转录调控机制,克隆其启动子,构建甘薯叶酵母单杂交cDNA文库,通过酵母单杂交方法筛选与IbHQT1启动子结合的上游调控因子。结果表明,1 500 bp的IbHQT1启动子序列中含有多种激素调节与防御相关的顺式元件及转录因子结合元件。构建的cDNA文库库容为1.15×107 CFU,插入片段长度平均约1 200 bp。通过酵母单杂交筛选获得2个转录因子IbMYB11和IbTGA2.2,可以与IbHQT1启动子结合。IbMYB11、IbTGA2.2和IbHQT1在甘薯(QS80-12-11)不同组织和发育阶段表达类似,与绿原酸的积累存在明显的相关性。
徐靖, 朱红林, 林延慧, 唐力琼, 唐清杰, 王效宁. 甘薯IbHQT1启动子的克隆及上游调控因子的鉴定[J]. 生物技术通报, 2023, 39(8): 213-219.
XU Jing, ZHU Hong-lin, LIN Yan-hui, TANG Li-qiong, TANG Qing-jie, WANG Xiao-ning. Cloning of IbHQT1 Promoter and Identification of Upstream Regulatory Factors in Sweet Potato[J]. Biotechnology Bulletin, 2023, 39(8): 213-219.
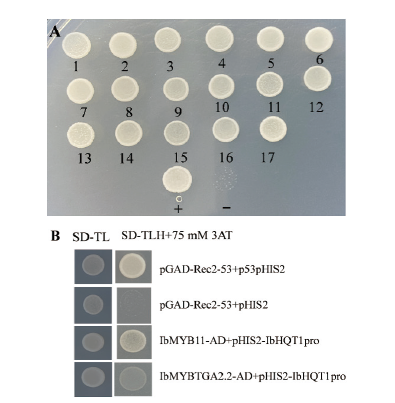
图4 IbHQT1启动子互作蛋白的筛选和鉴定 A:IbHQT1启动子结合蛋白的二次筛选,1-17:酵母菌落;+:阳性对照;-:阴性对照;B:酵母单杂交验证IbHQT1启动子与IbMYB11和IbTGA2.2的互作
Fig. 4 Screening and identification of IbHQT1 promoter interacting proteins A: Secondary screening of binding protein interacting with IbHQT1 promoter.1-17: Yeast colonies; +: positive control; -:negative control. B: The interaction of IbHQT1 promoter with IbMYB11 and IbTGA2.2
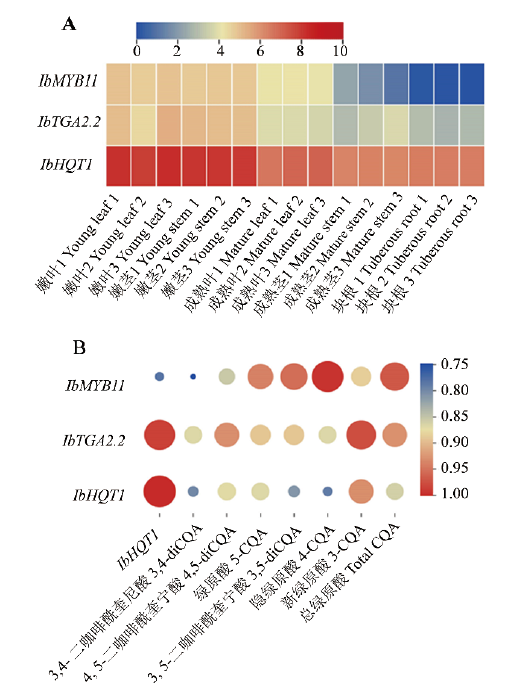
图5 IbMYB11、IbTGA2.2和IbHQT1共表达(A)及其与甘薯绿原酸积累相关性(B)
Fig. 5 Co-expression of IbMYB11, IbTGA2.2 and IbHQT1(A)and their correlations with chlorogenic acid accumulation in sweet potato(B)
| [1] |
Tajik N, Tajik M, Mack I, et al. The potential effects of chlorogenic acid, the main phenolic components in coffee, on health: a comprehensive review of the literature[J]. Eur J Nutr, 2017, 56(7): 2215-2244.
doi: 10.1007/s00394-017-1379-1 pmid: 28391515 |
| [2] |
Naveed M, Hejazi V, Abbas M, et al. Chlorogenic acid(CGA): A pharmacological review and call for further research[J]. Biomed Pharmacother, 2018, 97: 67-74.
doi: 10.1016/j.biopha.2017.10.064 URL |
| [3] |
Wianowska D, Gil M. Recent advances in extraction and analysis procedures of natural chlorogenic acids[J]. Phytochem Rev, 2019, 18(1): 273-302.
doi: 10.1007/s11101-018-9592-y |
| [4] |
Mohanraj R, Sivasankar S. Sweet potato(Ipomoea batatas[L.]Lam): a valuable medicinal food: a review[J]. J Med Food, 2014, 17(7): 733-741.
doi: 10.1089/jmf.2013.2818 pmid: 24921903 |
| [5] |
Tanaka M, Ishiguro K, Oki T, et al. Functional components in sweetpotato and their genetic improvement[J]. Breed Sci, 2017, 67(1): 52-61.
doi: 10.1270/jsbbs.16125 URL |
| [6] | Islam MS, Yoshimoto M, Yamakawa O. Distribution and physiological functions of caffeoylquinic acid derivatives in leaves of sweetpotato genotypes[J]. J Food Sci, 2006, 68(1): 111-116. |
| [7] |
Rautenbach F, Faber M, Laurie S, et al. Antioxidant capacity and antioxidant content in roots of 4 sweetpotato varieties[J]. J Food Sci, 2010, 75(5): C400-C405.
doi: 10.1111/jfds.2010.75.issue-5 URL |
| [8] |
李佳银, 于欢, 石伯阳, 等. 甘薯茎叶中异槲皮苷及咖啡酰基奎宁酸类衍生物的抗氧化活性[J]. 食品科学, 2013, 34(7): 111-114.
doi: 10.7506/spkx1002-6630-201307023 |
| Li JY, Yu H, Shi BY, et al. Antioxidant activity of isoquercitrin and caffeoylquinic acid derivatives from sweet potato stems and leaves[J]. Food sci, 2013, 34(7): 111-114. | |
| [9] | 刘雪辉, 李觅路, 谭斌, 等. 紫甘薯茎叶中绿原酸及异绿原酸对α-葡萄糖苷酶的抑制作用[J]. 现代食品科技, 2014, 30(3): 103-107. |
| Liu XH, Li ML, Tan B, et al. Inhibitory effects of chlorogenic acid and isochlorogenic acid from purple sweet potato leaves on α-glucosidase[J]. Mod Food Sci Technol, 2014, 30(3): 103-107. | |
| [10] | 王伟, 阮妙鸿, 邱永祥, 等. 甘薯抗薯瘟病的苯丙烷类代谢研究[J]. 中国生态农业学报, 2009, 17(5): 944-948. |
|
Wang W, Ruan MH, Qiu YX, et al. Phenylaprapanoid metabolism of sweet potato against Pseudomonas solanacearum[J]. Chin J Eco Agric, 2009, 17(5): 944-948.
doi: 10.3724/SP.J.1011.2009.00944 URL |
|
| [11] | 刘美艳, 孙厚俊, 王景景, 等. 甘薯块根抗黑斑病酚类物质代谢的研究[J]. 中国农学通报, 2012, 28(24): 226-230. |
| Liu MY, Sun HJ, Wang JJ, et al. The study on phenol metabolism in sweet potato infected by Ceratocystis fimbriata Ellis et Halsted[J]. Chin Agric Sci Bull, 2012, 28(24): 226-230. | |
| [12] |
刘海晶, 段立清, 李海平, 等. 绿原酸提高舞毒蛾核型多角体病毒(LdNPV)的致病力[J]. 昆虫学报, 2016, 59(5): 568-572.
doi: 10.16380/j.kcxb.2016.05.012 |
| Liu HJ, Duan LQ, Li HP, et al. Chlorogenic acid enhances the virulence of Lymantria dispar necleopolyhydrovirus(LdNPV)[J]. Acta Entomol Sin, 2016, 59(5): 568-572. | |
| [13] | 王珍, 李宗芸. 绿原酸的生物活性及甘薯绿原酸研究进展[J]. 江苏师范大学学报: 自然科学版, 2017, 35(3): 30-34, 48. |
| Wang Z, Li ZY. Research progress of biological activity of chlorogenic acid and chlorogenic acid in sweetpotato[J]. J Jiangsu Norm Univ Nat Sci Ed, 2017, 35(3): 30-34, 48. | |
| [14] |
Liu Y, Su WJ, Wang LJ, et al. Integrated transcriptome, small RNA and degradome sequencing approaches proffer insights into chlorogenic acid biosynthesis in leafy sweet potato[J]. PLoS One, 2021, 16(1): e0245266.
doi: 10.1371/journal.pone.0245266 URL |
| [15] |
Liu Q, Liu Y, Xu YC, et al. Overexpression of and RNA interference with hydroxycinnamoyl-CoA quinate hydroxycinnamoyl transferase affect the chlorogenic acid metabolic pathway and enhance salt tolerance in Taraxacum antungense Kitag[J]. Phytochem Lett, 2018, 28: 116-123.
doi: 10.1016/j.phytol.2018.10.003 URL |
| [16] |
Moglia A, Lanteri S, Comino C, et al. Dual catalytic activity of hydroxycinnamoyl-coenzyme A quinate transferase from tomato allows it to moonlight in the synthesis of both mono- and dicaffeoylquinic acids[J]. Plant Physiol, 2014, 166(4): 1777-1787.
doi: 10.1104/pp.114.251371 pmid: 25301886 |
| [17] |
Payyavula RS, Shakya R, Sengoda VG, et al. Synthesis and regulation of chlorogenic acid in potato: Rerouting phenylpropanoid flux in HQT-silenced lines[J]. Plant Biotechnol J, 2015, 13(4): 551-564.
doi: 10.1111/pbi.12280 pmid: 25421386 |
| [18] |
Zhang JR, Wu ML, Li WD, et al. Regulation of chlorogenic acid biosynthesis by hydroxycinnamoyl CoA quinate hydroxycinnamoyl transferase in Lonicera japonica[J]. Plant Physiol Biochem, 2017, 121: 74-79.
doi: 10.1016/j.plaphy.2017.10.017 URL |
| [19] | Moglia A, Acquadro A, Eljounaidi K, et al. Genome-wide identification of BAHD acyltransferases and in vivo characterization of HQT-like enzymes involved in caffeoylquinic acid synthesis in globe artichoke[J]. Front Plant Sci, 2016, 7: 1424. |
| [20] |
Mudau SP, Steenkamp PA, Piater LA, et al. Metabolomics-guided investigations of unintended effects of the expression of the hydroxycinnamoyl quinate hydroxycinnamoyltransferase(hqt1)gene from Cynara cardunculus var. scolymus in Nicotiana tabacum cell cultures[J]. Plant Physiol Biochem, 2018, 127: 287-298.
doi: 10.1016/j.plaphy.2018.04.005 URL |
| [21] |
Li YQ, Kong DX, Bai M, et al. Correlation of the temporal and spatial expression patterns of HQT with the biosynthesis and accumulation of chlorogenic acid in Lonicera japonica flowers[J]. Hortic Res, 2019, 6: 73.
doi: 10.1038/s41438-019-0154-2 |
| [22] |
Patra B, Schluttenhofer C, Wu YM, et al. Transcriptional regulation of secondary metabolite biosynthesis in plants[J]. Biochim Biophys Acta, 2013, 1829(11): 1236-1247.
doi: 10.1016/j.bbagrm.2013.09.006 pmid: 24113224 |
| [23] |
Chezem WR, Clay NK. Regulation of plant secondary metabolism and associated specialized cell development by MYBs and bHLHs[J]. Phytochemistry, 2016, 131: 26-43.
doi: S0031-9422(16)30158-3 pmid: 27569707 |
| [24] | 赵莹, 杨欣宇, 赵晓丹, 等. 植物类黄酮化合物生物合成调控研究进展[J]. 食品工业科技, 2021, 42(21): 454-463. |
| Zhao Y, Yang XY, Zhao XD, et al. Research progress on regulation of plant flavonoids biosynthesis[J]. Sci Technol Food Ind, 2021, 42(21): 454-463. | |
| [25] |
Xu J, Zhu JH, Lin YH, et al. Comparative transcriptome and weighted correlation network analyses reveal candidate genes involved in chlorogenic acid biosynthesis in sweet potato[J]. Sci Rep, 2022, 12(1): 1-11.
doi: 10.1038/s41598-021-99269-x |
| [26] |
Zhou ML, Memelink J. Jasmonate-responsive transcription factors regulating plant secondary metabolism[J]. Biotechnol Adv, 2016, 34(4): 441-449.
doi: S0734-9750(16)30011-8 pmid: 26876016 |
| [27] | 段乐鹏, 谢丽琼, 马艺沔, 等. 转录因子对药用植物次生代谢的调控作用[J]. 中药材, 2021, 44(4): 1002-1007. |
| Duan LP, Xie LQ, Ma YG, et al. Regulation of transcription factors on secondary metabolism of medicinal plants[J]. J Chin Med Mater, 2021, 44(4): 1002-1007. | |
| [28] |
Ma DW, Constabel CP. MYB repressors as regulators of phenylpropanoid metabolism in plants[J]. Trends Plant Sci, 2019, 24(3): 275-289
doi: S1360-1385(18)30290-5 pmid: 30704824 |
| [29] |
Anwar M, Chen L, Xiao YB, et al. Recent advanced metabolic and genetic engineering of phenylpropanoid biosynthetic pathways[J]. Int J Mol Sci, 2021, 22(17): 9544.
doi: 10.3390/ijms22179544 URL |
| [30] |
Duan AQ, Tan SS, Deng YJ, et al. Genome-wide identification and evolution analysis of R2R3-MYB gene family reveals S6 subfamily R2R3-MYB transcription factors involved in anthocyanin biosynthesis in carrot[J]. Int J Mol Sci, 2022, 23: 11859.
doi: 10.3390/ijms231911859 URL |
| [31] |
Zhao L, Song ZB, Wang BW, et al. R2R3-MYB transcription factor NtMYB330 regulates proanthocyanidin biosynthesis and seed germination in tobacco(Nicotiana tabacum L.)[J]. Front Plant Sci, 2022, 12: 819247.
doi: 10.3389/fpls.2021.819247 URL |
| [32] |
Tian Q, Han LM, Zhu XY, et al. SmMYB4 is a R2R3-MYB transcriptional repressor regulating the biosynthesis of phenolic acids and tanshinones in Salvia miltiorrhiza[J]. Metabolites, 2022, 12(10): 968.
doi: 10.3390/metabo12100968 URL |
| [33] |
Stracke R, Jahns O, Keck M, et al. Analysis of production of flavonol glycosides-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals myb11-, myb12- and myb111-independent flavonol glycoside accumulation[J]. New Phytol, 2010, 188(4): 985-1000.
doi: 10.1111/j.1469-8137.2010.03421.x pmid: 20731781 |
| [34] | Li Y, Chen M, Wang SL, et al. AtMYB11 regulates caffeoylquinic acid and flavonol synthesis in tomato and tobacco[J]. Plant Cell Tissue Organ Cult PCTOC, 2015, 122(2): 309-319. |
| [35] |
Luo J, Butelli E, Hill L, et al. AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: expression in fruit results in very high levels of both types of polyphenol[J]. Plant J, 2008, 56(2): 316-326.
doi: 10.1111/j.1365-313X.2008.03597.x URL |
| [36] |
Guo D, Li HL, Zhu JH, et al. HbTGA1, a TGA transcription factor from Hevea brasiliensis, regulates the expression of multiple natural rubber biosynthesis genes[J]. Front Plant Sci, 2022, 13: 909098.
doi: 10.3389/fpls.2022.909098 URL |
| [37] |
Huo YB, Zhang B, Chen L, et al. Isolation and functional characterization of the promoters of miltiradiene synthase genes, TwTPS27a and TwTPS27b, and interaction analysis with the transcription factor TwTGA1 from Tripterygium wilfordii[J]. Plants, 2021, 10(2): 418.
doi: 10.3390/plants10020418 URL |
| [38] |
Lyu ZY, Guo ZY, Zhang LD, et al. Interaction of BZIP transcription factor TGA6 with salicylic acid signaling modulates artemisinin biosynthesis in Artemisia annua[J]. J Exp Bot, 2019, 70(15): 3969-3979.
doi: 10.1093/jxb/erz166 URL |
| [39] |
Han J, Liu HT, Wang SC, et al. A class I TGA transcription factor from Tripterygium wilfordii Hook. f. modulates the biosynthesis of secondary metabolites in both native and heterologous hosts[J]. Plant Sci, 2020, 290: 110293.
doi: 10.1016/j.plantsci.2019.110293 URL |
| [40] |
Mao ZL, Jiang HY, Wang S, et al. The MdHY5-MdWRKY41-MdMYB transcription factor cascade regulates the anthocyanin and proanthocyanidin biosynthesis in red-fleshed apple[J]. Plant Sci, 2021, 306: 110848.
doi: 10.1016/j.plantsci.2021.110848 URL |
| [41] |
Sun QG, Jiang SH, Zhang TL, et al. Apple NAC transcription factor MdNAC52 regulates biosynthesis of anthocyanin and proanthocyanidin through MdMYB9 and MdMYB11[J]. Plant Sci, 2019, 289: 110286.
doi: 10.1016/j.plantsci.2019.110286 URL |
| [42] |
An JP, Yao JF, Xu RR, et al. Apple bZIP transcription factor MdbZIP44 regulates abscisic acid-promoted anthocyanin accumulation[J]. Plant Cell Environ, 2018, 41(11): 2678-2692.
doi: 10.1111/pce.v41.11 URL |
| [43] |
Xu WJ, Dubos C, Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes[J]. Trends Plant Sci, 2015, 20(3): 176-185.
doi: 10.1016/j.tplants.2014.12.001 pmid: 25577424 |
| [1] | 黄小龙, 孙贵连, 马丹丹, 闫慧清. 水稻幼苗酵母单杂文库构建及LAZY1上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 126-135. |
| [2] | 韩浩章, 张丽华, 李素华, 赵荣, 王芳, 王晓立. 盐碱胁迫诱导的猴樟酵母cDNA文库构建及CbP5CS上游调控因子筛选[J]. 生物技术通报, 2023, 39(9): 236-245. |
| [3] | 吕秋谕, 孙培媛, 冉彬, 王佳蕊, 陈庆富, 李洪有. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 194-203. |
| [4] | 李博, 刘合霞, 陈宇玲, 周兴文, 朱宇林. 金花茶CnbHLH79转录因子的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2023, 39(8): 241-250. |
| [5] | 陈晓, 于茗兰, 吴隆坤, 郑晓明, 逄洪波. 植物lncRNA及其对低温胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(7): 1-12. |
| [6] | 郭怡婷, 赵文菊, 任延靖, 赵孟良. 菊芋NAC转录因子家族基因的鉴定及分析[J]. 生物技术通报, 2023, 39(6): 217-232. |
| [7] | 冯珊珊, 王璐, 周益, 王幼平, 方玉洁. WOX家族基因调控植物生长发育和非生物胁迫响应的研究进展[J]. 生物技术通报, 2023, 39(5): 1-13. |
| [8] | 王兵, 赵会纳, 余婧, 余世洲, 雷波. 植物侧枝发育的调控研究进展[J]. 生物技术通报, 2023, 39(5): 14-22. |
| [9] | 张新博, 崔浩亮, 史佩华, 高锦春, 赵顺然, 陶晨雨. 低起始量的免疫共沉淀技术研究进展[J]. 生物技术通报, 2023, 39(4): 227-235. |
| [10] | 葛颜锐, 赵冉, 徐静, 李若凡, 胡云涛, 李瑞丽. 植物维管形成层发育及其调控的研究进展[J]. 生物技术通报, 2023, 39(3): 13-25. |
| [11] | 刘铖霞, 孙宗艳, 罗云波, 朱鸿亮, 曲桂芹. bHLH转录因子的磷酸化调控植物生理功能的研究进展[J]. 生物技术通报, 2023, 39(3): 26-34. |
| [12] | 赵孟良, 郭怡婷, 任延靖. 菊芋WRKY转录因子家族基因的鉴定及分析[J]. 生物技术通报, 2023, 39(2): 116-125. |
| [13] | 韩芳英, 胡昕, 王楠楠, 谢裕红, 王晓艳, 朱强. DREBs响应植物非生物逆境胁迫研究进展[J]. 生物技术通报, 2023, 39(11): 86-98. |
| [14] | 陈楚怡, 杨小梅, 陈胜艳, 陈斌, 岳莉然. ABA和干旱胁迫下菊花脑ZF-HD基因家族的表达分析[J]. 生物技术通报, 2023, 39(11): 270-282. |
| [15] | 冯策婷, 江律, 刘鑫颖, 罗乐, 潘会堂, 张启翔, 于超. 单叶蔷薇NAC基因家族鉴定及干旱胁迫响应分析[J]. 生物技术通报, 2023, 39(11): 283-296. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||