








生物技术通报 ›› 2024, Vol. 40 ›› Issue (6): 203-218.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0907
王健1,2( ), 杨莎1,2, 孙庆文1,2, 陈宏宇1,2, 杨涛1, 黄园1,2(
), 杨莎1,2, 孙庆文1,2, 陈宏宇1,2, 杨涛1, 黄园1,2( )
)
收稿日期:2023-09-21
出版日期:2024-06-26
发布日期:2024-06-24
通讯作者:
黄园,女,博士,讲师,研究方向:中药资源与开发;E-mail: huangyuanhy2014@163.com作者简介:王健,男,硕士研究生,研究方向:中药资源与开发;E-mail: 3078048759@qq.com
基金资助:
WANG Jian1,2( ), YANG Sha1,2, SUN Qing-wen1,2, CHEN Hong-yu1,2, YANG Tao1, HUANG Yuan1,2(
), YANG Sha1,2, SUN Qing-wen1,2, CHEN Hong-yu1,2, YANG Tao1, HUANG Yuan1,2( )
)
Received:2023-09-21
Published:2024-06-26
Online:2024-06-24
摘要:
【目的】 bHLH转录因子广泛存在于植物中,是调控植物生长发育和响应逆境胁迫的重要转录因子家族之一。从金钗石斛全基因组中鉴定DnbHLHs转录因子家族,分析其表达特点,为后续深入研究DnbHLHs转录因子的生物学功能奠定基础。【方法】 利用生物信息学方法从金钗石斛全基因组中鉴定出bHLH转录因子家族成员,并对其理化特征、系统进化关系、基因结构、染色体定位和启动子顺式作用元件等进行分析,利用转录组数据和RT-qPCR分析DnbHLHs转录因子的组织表达模式和不同非生物胁迫下的表达规律。【结果】 在金钗石斛中共鉴定出137个bHLH转录因子,DnbHLHs蛋白的氨基酸数量为85-1 260,分子量为9 855.23-140 024.19 Da,等电点为4.52-10.66,不稳定系数为31.94-87.42,脂溶指数为56.66-106.06,亲水指数为-0.815-0.185。DnbHLHs分布于金钗石斛的19条染色体上,都存在bHLH结构域,与拟南芥的bHLH同源,且组内、种间存在着共线性关系。基因表达模式分析显示,DnbHLHs家族成员存在组织表达特异性,且DnbHLH26和DnbHLH33对多种逆境胁迫有响应。【结论】 从金钗石斛全基因组中鉴定出137个DnbHLHs转录因子家族成员,该家族成员的理化性质和保守基序存在特异性和多样性的特征,不同DnbHLHs转录因子之间具有组织表达差异性,且DnbHLH26主要受高盐和高温胁迫的诱导,DnbHLH33主要受高盐、高温和低温胁迫的诱导。
王健, 杨莎, 孙庆文, 陈宏宇, 杨涛, 黄园. 金钗石斛bHLH转录因子家族全基因组鉴定及表达分析[J]. 生物技术通报, 2024, 40(6): 203-218.
WANG Jian, YANG Sha, SUN Qing-wen, CHEN Hong-yu, YANG Tao, HUANG Yuan. Genome-wide Identification and Expression Analysis of bHLH Transcription Factor Family in Dendrobium nobile[J]. Biotechnology Bulletin, 2024, 40(6): 203-218.
| 基因 Gene | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|
| DnbHLH26 | GGTCCTTCGCCATCTTC | CGTCAGTAACTCGGTCAA |
| DnbHLH33 | ACGCAGTCCTTCATCAAC | GTTCATCATCTCCACACTATG |
| Ubiquitin | CGCCGTCAACCTCATTCCAT | GTGAGGTAGCGACCGTGGC |
表1 引物序列
Table 1 Primer sequences
| 基因 Gene | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|
| DnbHLH26 | GGTCCTTCGCCATCTTC | CGTCAGTAACTCGGTCAA |
| DnbHLH33 | ACGCAGTCCTTCATCAAC | GTTCATCATCTCCACACTATG |
| Ubiquitin | CGCCGTCAACCTCATTCCAT | GTGAGGTAGCGACCGTGGC |
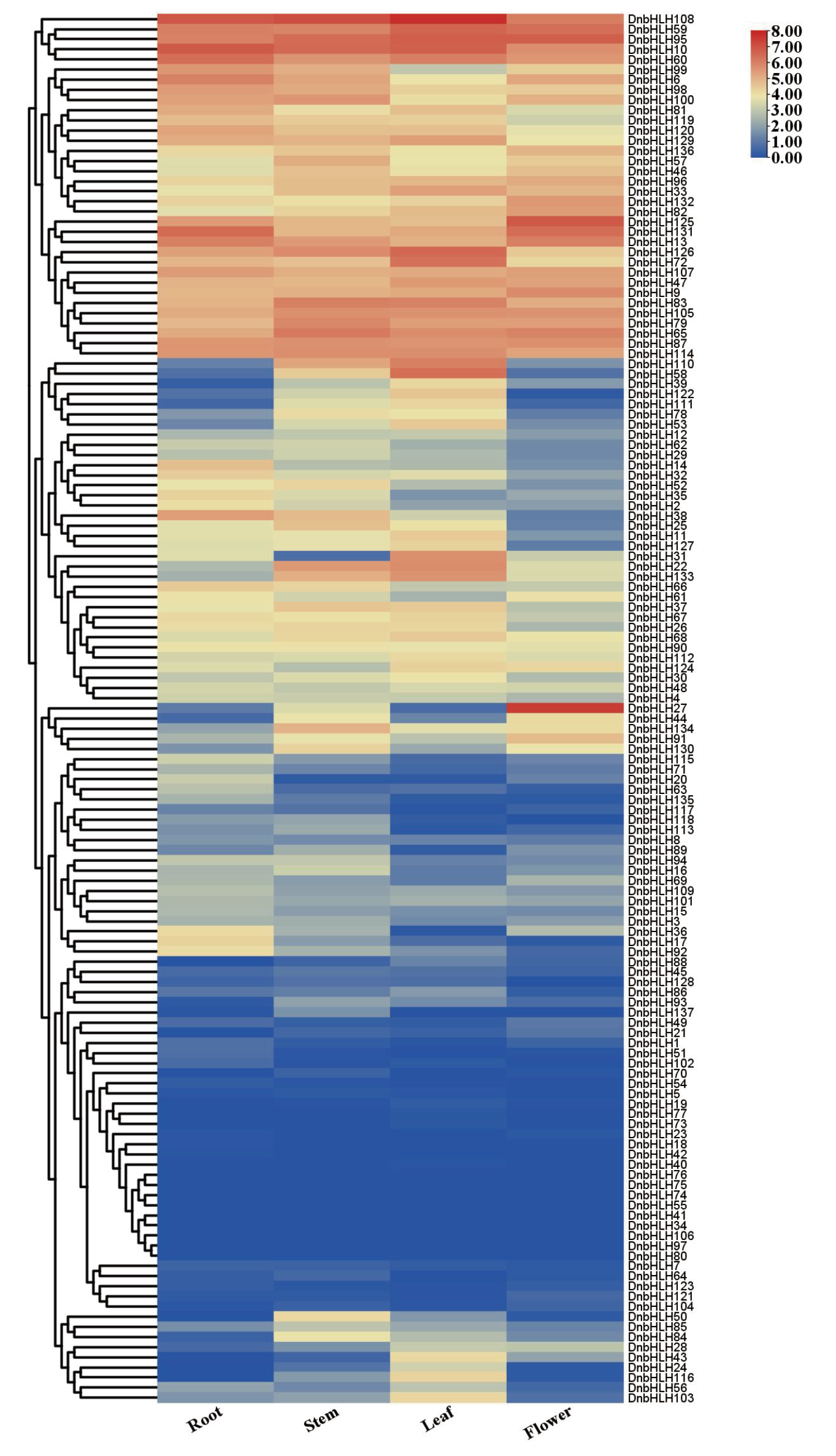
图10 金钗石斛bHLH转录因子家族表达模式 色阶表示Log2(TPM+1)的值
Fig. 10 Expression patterns of bHLH transcription factor family in D. nobile The level represents the value of Log2(TPM+1)
| [1] |
Waseem M, Nkurikiyimfura O, Niyitanga S, et al. GRAS transcription factors emerging regulator in plants growth, development, and multiple stresses[J]. Mol Biol Rep, 2022, 49(10): 9673-9685.
doi: 10.1007/s11033-022-07425-x pmid: 35713799 |
| [2] | 令狐楚, 谷荣辉, 秦礼康. 金钗石斛的化学成分及药理作用研究进展[J]. 中草药, 2021, 52(24): 7693-7708. |
| Linghu C, Gu RH, Qin LK. Research progress on chemical constituents and pharmacological effects of Dendrobium nobile[J]. Chin Tradit Herb Drugs, 2021, 52(24): 7693-7708. | |
| [3] | Zheng SG, Hu YD, Zhao RX, et al. Quantitative assessment of secondary metabolites and cancer cell inhibiting activity by high performance liquid chromatography fingerprinting in Dendrobium nobile[J]. J Chromatogr B Analyt Technol Biomed Life Sci, 2020, 1140: 122017. |
| [4] | Luo AX, He XJ, Zhou SD, et al. Purification, composition analysis and antioxidant activity of the polysaccharides from Dendrobium nobile Lindl[J]. Carbohydr Polym, 2010, 79(4): 1014-1019. |
| [5] | Nie XQ, Chen Y, Li W, et al. Anti-aging properties of Dendrobium nobile Lindl.: from molecular mechanisms to potential treatments[J]. J Ethnopharmacol, 2020, 257: 112839. |
| [6] | 张明, 夏鸿西, 朱利泉. 石斛组织培养研究进展[J]. 中国中药杂志, 2000, 25(6): 323. |
| Zhang M, Xia HX, Zhu LQ. Research progress of Dendrobium tissue culture[J]. China J Chin Mater Med, 2000, 25(6): 323. | |
| [7] | Guo AY, Chen X, Gao G, et al. PlantTFDB: a comprehensive plant transcription factor database[J]. Nucleic Acids Res, 2008, 36(Database issue): D966-D969. |
| [8] |
Atchley WR, Terhalle W, Dress A. Positional dependence, cliques, and predictive motifs in the bHLH protein domain[J]. J Mol Evol, 1999, 48(5): 501-516.
doi: 10.1007/pl00006494 pmid: 10198117 |
| [9] |
Albert NW, Lewis DH, Zhang HB, et al. Light-induced vegetative anthocyanin pigmentation in Petunia[J]. J Exp Bot, 2009, 60(7): 2191-2202.
doi: 10.1093/jxb/erp097 pmid: 19380423 |
| [10] |
Lu J, Webb R, Richardson JA, et al. MyoR: a muscle-restricted basic helix-loop-helix transcription factor that antagonizes the actions of MyoD[J]. Proc Natl Acad Sci USA, 1999, 96(2): 552-557.
pmid: 9892671 |
| [11] |
Heim MA, Jakoby M, Werber M, et al. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity[J]. Mol Biol Evol, 2003, 20(5): 735-747.
doi: 10.1093/molbev/msg088 pmid: 12679534 |
| [12] | 李洁. 植物转录因子与基因调控[J]. 生物学通报, 2004, 39(3): 9-11. |
| Li J. Plant transcription factors and gene regulation[J]. Bull Biol, 2004, 39(3): 9-11. | |
| [13] | Seo JS, Joo J, Kim MJ, et al. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice[J]. Plant J, 2011, 65(6): 907-921. |
| [14] | 王玉军, 郝宇钧, 戴继勋, 等. OsbHLH1基因在拟南芥中表达及耐低温能力的研究[J]. 高技术通讯, 2004, 14(4): 35-38. |
| Wang YJ, Hao YJ, Dai JX, et al. Expression and low temperature tolerance of OsbHLH1 gene in Arabidopsis thaliana[J]. High Technology Communication, 2004, 14(4): 4-12. | |
| [15] | Zhang J, Liu B, Li MS, et al. The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis[J]. Plant Cell, 2015, 27(3): 787-805. |
| [16] |
Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [17] | Cui BL, Huang M, Guo CD, et al. Cloning and expression analysis of DnMSI1 gene in orchid species Dendrobium nobile Lindl[J]. Plant Signal Behav, 2022, 17(1): 2021649. |
| [18] | 郝立冬, 郭海滨, 李明, 等. 玉米MYC基因家族的结构特点和表达模式分析[J]. 基因组学与应用生物学, 2021, 40(4): 1748-1753. |
| Hao LD, Guo HB, Li M, et al. Analysis of characteristics and expression pattern of maize MYC gene family[J]. Genom Appl Biol, 2021, 40(4): 1748-1753. | |
| [19] | 王力伟, 房永雨, 刘红葵, 等. bHLH转录因子的研究进展[J]. 畜牧与饲料科学, 2020, 41(1): 23-27. |
| Wang LW, Fang YY, Liu HK, et al. Research progress of bHLH transcription factors[J]. Anim Husb Feed Sci, 2020, 41(1): 23-27. | |
| [20] | 何开平, 吴楚. bHLH转录因子对植物形态发生的影响[J]. 安徽农业科学, 2010, 38(35): 19957-19959. |
| He KP, Wu C. The effects of bHLH transcription factors on plant morphogenesis[J]. J Anhui Agric Sci, 2010, 38(35): 19957-19959. | |
| [21] | Zhang TT, Lv W, Zhang HS, et al. Genome-wide analysis of the basic Helix-Loop-Helix(bHLH)transcription factor family in maize[J]. BMC Plant Biol, 2018, 18(1): 235. |
| [22] | Wei KF, Chen HQ. Comparative functional genomics analysis of bHLH gene family in rice, maize and wheat[J]. BMC Plant Biol, 2018, 18(1): 309. |
| [23] | 徐秀荣, 杨克彬, 王思宁, 等. 毛竹bHLH转录因子的鉴定及其在干旱和盐胁迫条件下的表达分析[J]. 植物科学学报, 2019, 37(5): 610-620. |
| Xu XR, Yang KB, Wang SN, et al. Identification of bHLH transcription factors in moso bamboo(Phyllostachys edulis)and their expression analysis under drought and salt stress[J]. Plant Sci J, 2019, 37(5): 610-620. | |
| [24] | Wang Y, Liu AZ. Genomic characterization and expression analysis of basic Helix-loop-Helix(bHLH)family genes in traditional Chinese herb Dendrobium officinale[J]. Plants, 2020, 9(8): 1044. |
| [25] | Wei S, Xin C, Hui L, et al. Genome-wide identification and analyses of bHLH family genes in Brassica napus[J]. Canadian Journal of Plant Science, 2019, 99(5): 589-598. |
| [26] |
Kavas M, Baloğlu MC, Atabay ES, et al. Genome-wide characterization and expression analysis of common bean bHLH transcription factors in response to excess salt concentration[J]. Mol Genet Genomics, 2016, 291(1): 129-143.
doi: 10.1007/s00438-015-1095-6 pmid: 26193947 |
| [27] |
Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family[J]. Plant Cell, 2003, 15(8): 1749-1770.
doi: 10.1105/tpc.013839 pmid: 12897250 |
| [28] | 武家颂, 崔欣茹, 张景荣, 等. 拟南芥bHLH转录因子在根毛发育中的作用[J]. 杭州师范大学学报: 自然科学版, 2022, 21(2): 144-151. |
| Wu JS, Cui XR, Zhang JR, et al. Role of bHLH transcription factor members in Arabidopsis root hair development[J]. J Hangzhou Norm Univ Nat Sci Ed, 2022, 21(2): 144-151. | |
| [29] | Liu WW, Tai HH, Li SS, et al. bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism[J]. New Phytol, 2014, 201(4): 1192-1204. |
| [30] |
刘文文, 李文学. 植物bHLH转录因子研究进展[J]. 生物技术进展, 2013, 3(1): 7-11.
doi: 10.3969/j.issn.2095-2341.2013.01.02 |
| Liu WW, Li WX. Progress of plant bHLH transcription factor[J]. Curr Biotechnol, 2013, 3(1): 7-11. | |
| [31] |
冯建英, 李立芹, 鲁黎明. 马铃薯bHLH转录因子家族全基因组鉴定与表达分析[J]. 生物技术通报, 2022, 38(2): 21-33.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-1327 |
| Feng JY, Li LQ, Lu LM. Genome-wide identification and expression analysis of the bHLH transcription factor family in Solanum tuberosum[J]. Biotechnol Bull, 2022, 38(2): 21-33. | |
| [32] |
Pires N, Dolan L. Origin and diversification of basic-helix-loop-helix proteins in plants[J]. Mol Biol Evol, 2010, 27(4): 862-874.
doi: 10.1093/molbev/msp288 pmid: 19942615 |
| [33] |
高莉娟, 张正社, 文裕, 等. 象草全基因组bHLH转录因子家族鉴定及表达分析[J]. 草业学报, 2022, 31(3): 47-59.
doi: 10.11686/cyxb2020593 |
| Gao LJ, Zhang ZS, Wen Y, et al. Genome-wide identification and expression analysis of the bHLH transcription factor family in Cenchrus purpureus[J]. Acta Prataculturae Sin, 2022, 31(3): 47-59. | |
| [34] | 朱涛, 李芳菲, 杨海涵, 等. 山药bHLH基因家族鉴定及表达分析[J]. 信阳师范学院学报: 自然科学版, 2022, 35(3): 393-399. |
| Zhu T, Li FF, Yang HH, et al. Identification and expression ananlysis of bHLH gene family in Chinese yam(Dioscoreae rhizoma)[J]. J Xinyang Norm Univ Nat Sci Ed, 2022, 35(3): 393-399. | |
| [35] | 廖桂花, 段钰, 王丛丛, 等. 赪桐bHLH转录因子家族鉴定及生物信息学分析[J/OL]. 分子植物育种, 2023: 1-17. http://kns.cnki.net/kcms/detail/46.1068.S.20230529.1303.022.html. |
| Liao GH, Duan Y, Wang CC, et al. Clerod bHLH transcription factor family identification and bioinformatics analysis[J/OL]. Molecular Plant Breeding: 1-17. http://kns.cnki.net/kcms/detail/46.1068.S.20230529.1303.022.html. | |
| [36] | Wang PF, Wang HL, Wang YM, et al. Analysis of bHLH genes from foxtail millet(Setaria italica)and their potential relevance to drought stress[J]. PLoS One, 2018, 13(11): e0207344. |
| [37] | Fan Y, Lai DL, Yang H, et al. Genome-wide identification and expression analysis of the bHLH transcription factor family and its response to abiotic stress in foxtail millet(Setaria italica L.)[J]. BMC Genomics, 2021, 22(1): 778. |
| [38] |
Zhan H, Liu HZ, Ai WF, et al. Genome-wide identification and expression analysis of the bHLH transcription factor family and its response to abiotic stress in Mongolian oak(Quercus mongolica)[J]. Curr Issues Mol Biol, 2023, 45(2): 1127-1148.
doi: 10.3390/cimb45020075 pmid: 36826020 |
| [39] | Muhammad H K, Xuemei F, Huabo W, et al. Genome-wide identification and expression analysis of the bHLH transcription factor family in wintersweet(Chimonanthus praecox)[J]. International Journal of Molecular Sciences, 2023, 24(17): 13461. |
| [40] | 陈媞颖, 刘娟, 袁媛, 等. 黄芩bHLH转录因子基因家族生物信息学及表达分析[J]. 中草药, 2018, 49(3): 671-677. |
| Chen TY, Liu J, Yuan Y, et al. Analysis of bioinformatics and expression level of bHLH transcription factors in Scutellaria baicalensis[J]. Chin Tradit Herb Drugs, 2018, 49(3): 671-677. | |
| [41] | 何昌文, 朱丽, 沈珊, 等. 银杏bHLH91转录因子基因的克隆及表达分析[J]. 广西植物, 2018, 38(2): 202-209. |
| He CW, Zhu L, Shen S, et al. Cloning and expression analysis of a bHLH91 transcription factor gene from Ginkgo biloba[J]. Guihaia, 2018, 38(2): 202-209. | |
| [42] | 孟富宣, 杨玉皎, 段元杰, 等. 甜橙bHLH转录因子家族的鉴定及生物信息学分析[J]. 西南林业大学学报: 自然科学, 2020, 40(5): 73-86. |
| Meng FX, Yang YJ, Duan YJ, et al. Identification and bioinformatics analysis of the bHLH transcription factor family in Citrus sinensis[J]. J Southwest For Univ Nat Sci, 2020, 40(5): 73-86. | |
| [43] | Guo JR, Sun BX, He HR, et al. Current understanding of bHLH transcription factors in plant abiotic stress tolerance[J]. Int J Mol Sci, 2021, 22(9): 4921. |
| [44] | Chinnusamy V, Ohta M, Kanrar S, et al. ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis[J]. Genes Dev, 2003, 17(8): 1043-1054. |
| [1] | 张迪, 鞠睿, 李丽梅, 王煜倩, 陈瑞, 王新一. 基于转录因子生物传感器在环境分析中的应用[J]. 生物技术通报, 2024, 40(6): 114-125. |
| [2] | 胡雅丹, 伍国强, 刘晨, 魏明. MYB转录因子在调控植物响应逆境胁迫中的作用[J]. 生物技术通报, 2024, 40(6): 5-22. |
| [3] | 胡永波, 雷雨田, 杨永森, 陈馨, 林黄昉, 林碧英, 刘爽, 毕格, 申宝营. 黄瓜和南瓜Bcl-2相关抗凋亡家族全基因组鉴定与表达模式分析[J]. 生物技术通报, 2024, 40(6): 219-237. |
| [4] | 王秋月, 段鹏亮, 李海笑, 刘宁, 曹志艳, 董金皋. 玉米大斑病菌cDNA文库的构建及转录因子StMR1互作蛋白的筛选[J]. 生物技术通报, 2024, 40(6): 281-289. |
| [5] | 常雪瑞, 王田田, 王静. 辣椒E2基因家族的鉴定及分析[J]. 生物技术通报, 2024, 40(6): 238-250. |
| [6] | 刘蓉, 田闵玉, 李光泽, 谭成方, 阮颖, 刘春林. 甘蓝型油菜REVEILLE家族鉴定及诱导表达分析[J]. 生物技术通报, 2024, 40(6): 161-171. |
| [7] | 王迪, 张晓宇, 宋宇鑫, 郑东然, 田静, 李玉花, 王宇, 吴昊. 细胞全能性转录因子调控植物组培再生的分子机制研究进展[J]. 生物技术通报, 2024, 40(6): 23-33. |
| [8] | 李梦然, 叶伟, 李赛妮, 张维阳, 李建军, 章卫民. Lithocarols类化合物生物合成基因litI的表达及其启动子功能分析[J]. 生物技术通报, 2024, 40(6): 310-318. |
| [9] | 杜兵帅, 邹昕蕙, 王子豪, 张馨元, 曹一博, 张凌云. 油茶SWEET基因家族的全基因组鉴定及表达分析[J]. 生物技术通报, 2024, 40(5): 179-190. |
| [10] | 侯雅琼, 郎红珊, 闻蒙蒙, 谷易云, 朱润洁, 汤晓丽. 猕猴桃AcHSP20基因家族的鉴定及表达分析[J]. 生物技术通报, 2024, 40(5): 167-178. |
| [11] | 李嘉欣, 李鸿燕, 刘丽娥, 张恬, 周武. 沙棘NRAMP基因家族鉴定及铅胁迫下表达分析[J]. 生物技术通报, 2024, 40(5): 191-202. |
| [12] | 李慧, 文钰芳, 王悦, 纪超, 石国优, 罗英, 周勇, 李志敏, 吴晓玉, 杨有新, 刘建萍. 盐胁迫下辣椒CaPIF4的表达特性与功能分析[J]. 生物技术通报, 2024, 40(4): 148-158. |
| [13] | 肖雅茹, 贾婷婷, 罗丹, 武喆, 李丽霞. 黄瓜CsERF025L转录因子的克隆及表达分析[J]. 生物技术通报, 2024, 40(4): 159-166. |
| [14] | 郭纯, 宋桂梅, 闫艳, 邸鹏, 王英平. 西洋参bZIP基因家族全基因组鉴定和表达分析[J]. 生物技术通报, 2024, 40(4): 167-178. |
| [15] | 钟匀, 林春, 刘正杰, 董陈文华, 毛自朝, 李兴玉. 芦笋皂苷合成相关糖基转移酶基因克隆及原核表达分析[J]. 生物技术通报, 2024, 40(4): 255-263. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||