








生物技术通报 ›› 2024, Vol. 40 ›› Issue (6): 190-202.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0012
收稿日期:2024-01-04
出版日期:2024-06-26
发布日期:2024-06-24
通讯作者:
高飞,男,教授,研究方向:生物化学与分子生物学;E-mail: gaofei@muc.edu.cn;作者简介:李博静,女,硕士研究生,研究方向:生物化学与分子生物学;E-mail: c2236363710@163.com
基金资助:
LI Bo-jing( ), ZHENG La-mei, WU Wu-yun, GAO Fei(
), ZHENG La-mei, WU Wu-yun, GAO Fei( ), ZHOU Yi-jun(
), ZHOU Yi-jun( )
)
Received:2024-01-04
Published:2024-06-26
Online:2024-06-24
摘要:
【目的】 研究西蒙得木HSP20家族成员的结构和功能,为后续研究提供支撑。【方法】 从结构分析、系统发育、表达模式等方面对西蒙得木HSP20家族成员(ScHSP20s)进行系统分析,并对ScHSP20-17进行功能分析。【结果】 西蒙得木基因组中包含35个HSP20基因座位,其中8个基因涉及串联重复,5个涉及片段重复。系统发育分析表明,35个ScHSP20聚为12个亚家族,其中核质亚家族数量最多。启动子分析表明,多数ScHSP20家族成员启动子区域含有激素响应和非生物胁迫等相关顺式作用元件。转录组分析表明,ScHSP20-35和ScHSP20-17等部分热激蛋白基因在花和种子发育过程中表达量发生显著性改变,而ScHSP20-17和ScHSP20-28等基因受高温诱导。亚细胞定位分析表明,ScHSP20-17编码的蛋白定位于细胞核中,并且在大肠杆菌、酵母和烟草中过表达ScHSP20-17能显著提高它们对高温胁迫的耐受性。【结论】 部分ScHSP20家族成员参与了西蒙得木生殖发育和非生物胁迫应答。
李博静, 郑腊梅, 吴乌云, 高飞, 周宜君. 西蒙得木HSP20基因家族的进化、表达和功能分析[J]. 生物技术通报, 2024, 40(6): 190-202.
LI Bo-jing, ZHENG La-mei, WU Wu-yun, GAO Fei, ZHOU Yi-jun. Evolution, Expression, and Functional Analysis of the HSP20 Gene Family from Simmondisa chinensis[J]. Biotechnology Bulletin, 2024, 40(6): 190-202.
| 基因名称Gene name | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|
| 18S | cgttaacgaacgagacctca | cccagaacatctaagggcat |
| ScHSP20-1 | gttgcggtcgcatttgttg | cgtccacggaagccatagaa |
| ScHSP20-3 | gccctgaaaatgaagagggtac | cctttgaacttggcttggatg |
| ScHSP20-4 | aatcaaaggagaaggggagaaa | aggaaccaccaccttcaacact |
| ScHSP20-13 | cgtcaagagggactcaccaaa | caccagaaaccgaaggagga |
| ScHSP20-14 | tcaagagggactcaccaaacag | agccacgtaagccccaatc |
| ScHSP20-16 | acacccttcatcacatcctcg | agctgttggggtactccttcac |
| ScHSP20-17 | accgtgtagagcggagcagt | cagtgacagtcaggaccccat |
| ScHSP20-20 | cagtaagagggcggaggatg | aagggaaagacgacgaggc |
| ScHSP20-25 | atggcgagcaaggcgttgacatgca | tgcatgtcaacgccttgctcgccat |
| ScHSP20-28 | tggcctctgtacgtgctgatt | caccctgttctcctccacttct |
| ScHSP20-23 | tgaggaacctccgacaaagac | cctcctgatgttgacgatgaaa |
| ScHSP20-26 | tccaggtcttagcaaggagca | tgtctttccctgtgggttcg |
| ScHSP20-32 | ctccctctgactgggtttcg | gggcgttctccacttatcctta |
表1 荧光定量PCR引物
Table 1 Primers used in RT-qPCR
| 基因名称Gene name | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|
| 18S | cgttaacgaacgagacctca | cccagaacatctaagggcat |
| ScHSP20-1 | gttgcggtcgcatttgttg | cgtccacggaagccatagaa |
| ScHSP20-3 | gccctgaaaatgaagagggtac | cctttgaacttggcttggatg |
| ScHSP20-4 | aatcaaaggagaaggggagaaa | aggaaccaccaccttcaacact |
| ScHSP20-13 | cgtcaagagggactcaccaaa | caccagaaaccgaaggagga |
| ScHSP20-14 | tcaagagggactcaccaaacag | agccacgtaagccccaatc |
| ScHSP20-16 | acacccttcatcacatcctcg | agctgttggggtactccttcac |
| ScHSP20-17 | accgtgtagagcggagcagt | cagtgacagtcaggaccccat |
| ScHSP20-20 | cagtaagagggcggaggatg | aagggaaagacgacgaggc |
| ScHSP20-25 | atggcgagcaaggcgttgacatgca | tgcatgtcaacgccttgctcgccat |
| ScHSP20-28 | tggcctctgtacgtgctgatt | caccctgttctcctccacttct |
| ScHSP20-23 | tgaggaacctccgacaaagac | cctcctgatgttgacgatgaaa |
| ScHSP20-26 | tccaggtcttagcaaggagca | tgtctttccctgtgggttcg |
| ScHSP20-32 | ctccctctgactgggtttcg | gggcgttctccacttatcctta |
| 用途 Purpose | 基因名称 Gene name | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|---|
| 原核表达载体 Prokaryotic expression vector | ScHSP20-17 | cgcgtggatccccggATGTCGCTTATTCCAAGTAGGT | agtcagtcacgatgcACCAGAGATTTCAATGGCTTTG |
| 酵母表达载体 Yeast expression vector | ScHSP20-17 | ctatagggaatattaATGTCGCTTATTCCAAGTAGGT | cggccgttactagtgACCAGAGATTTCAATGGCTTTG |
| 亚细胞定位载体 Subcellular localization vector | ScHSP20-17 | agtccggagctagctATGTCGCTTATTCCAAGTAGGT | cccttgctcaccatgACCAGAGATTTCAATGGCTTTG |
| 内参基因 Internal reference gene | Actin3 | GAGGGCCGTGTTCCCCAGCATCGTC | TCTTTTTGATTGAGCCTCATCCCCT |
| 基因表达分析 Gene expression analysis | ScHSP20-17 | AAGAATGATACCTGGCACCG | TGACCTCCTCCTTAGGCACA |
表2 引物序列
Table 2 Primer sequences
| 用途 Purpose | 基因名称 Gene name | 上游引物 Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|---|
| 原核表达载体 Prokaryotic expression vector | ScHSP20-17 | cgcgtggatccccggATGTCGCTTATTCCAAGTAGGT | agtcagtcacgatgcACCAGAGATTTCAATGGCTTTG |
| 酵母表达载体 Yeast expression vector | ScHSP20-17 | ctatagggaatattaATGTCGCTTATTCCAAGTAGGT | cggccgttactagtgACCAGAGATTTCAATGGCTTTG |
| 亚细胞定位载体 Subcellular localization vector | ScHSP20-17 | agtccggagctagctATGTCGCTTATTCCAAGTAGGT | cccttgctcaccatgACCAGAGATTTCAATGGCTTTG |
| 内参基因 Internal reference gene | Actin3 | GAGGGCCGTGTTCCCCAGCATCGTC | TCTTTTTGATTGAGCCTCATCCCCT |
| 基因表达分析 Gene expression analysis | ScHSP20-17 | AAGAATGATACCTGGCACCG | TGACCTCCTCCTTAGGCACA |
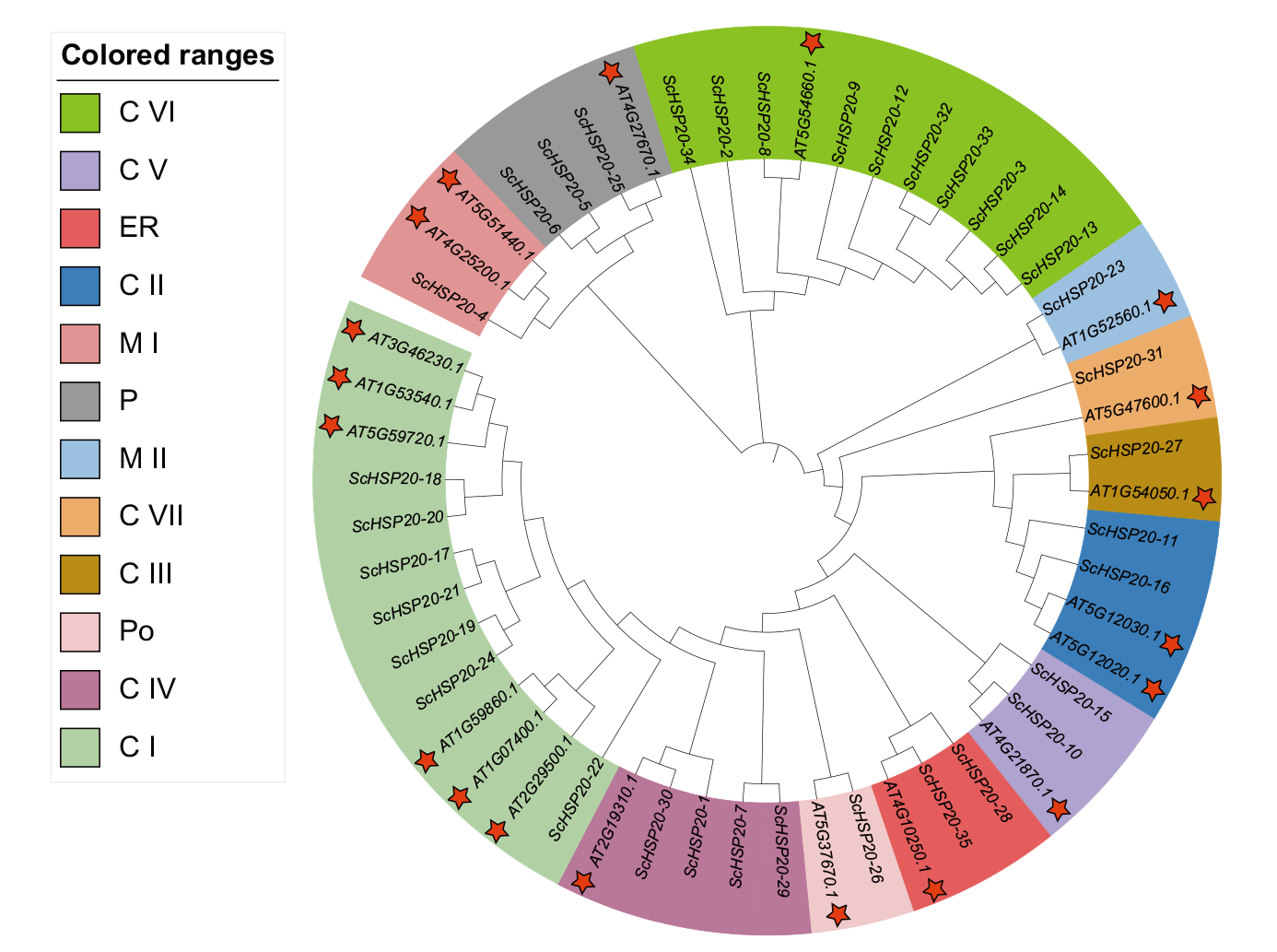
图3 拟南芥和西蒙得木HSP20基因的进化分析 C I、C II、C III、C IV、C V、C VI、C VII:细胞质或细胞核;MI、MII:线粒体;ER:内质网;P:质体;Po:过氧化物酶体
Fig. 3 Evolutionary analysis of the HSP20 gene in Arabidopsis and S. chinensis C I, C II, C III, CIV, C V, C VI, C VII: Cytoplasm or nucleus. Ml, MII: Mitochondria. ER: Endoplasmic reticulum. P: Plastid. Po: Peroxisome
| 重复基因对 Repeated gene pairs | Ka | Ks | Ka/Ks |
|---|---|---|---|
| ScHSP20-2 vs ScHSP20-17 | 0.383 89 | 2.622 38 | 0.146 39 |
| ScHSP20-5 vs ScHSP20-6 | 0.003 77 | 0.027 34 | 0.137 86 |
| ScHSP20-17 vs ScHSP20-24 | 0.137 24 | 1.495 32 | 0.091 78 |
表3 ScHSP20家族成员中片段重复基因对的Ka/Ks值
Table 3 Ka/Ks values of fragment repeat gene pairs in ScHSP20 family members
| 重复基因对 Repeated gene pairs | Ka | Ks | Ka/Ks |
|---|---|---|---|
| ScHSP20-2 vs ScHSP20-17 | 0.383 89 | 2.622 38 | 0.146 39 |
| ScHSP20-5 vs ScHSP20-6 | 0.003 77 | 0.027 34 | 0.137 86 |
| ScHSP20-17 vs ScHSP20-24 | 0.137 24 | 1.495 32 | 0.091 78 |
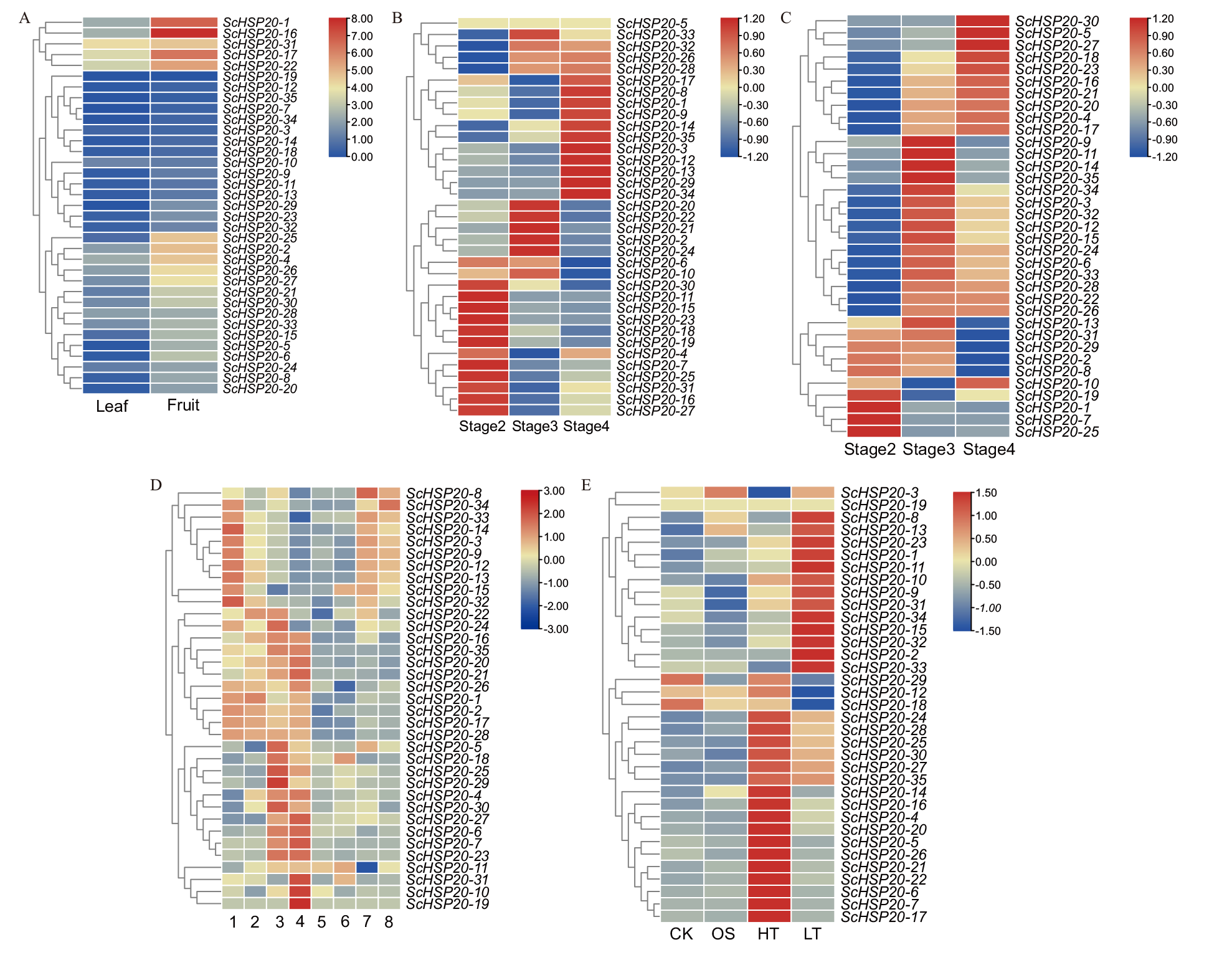
图5 ScHSP20基因在不同组织及不同胁迫下的表达 A:叶片和果实;B:雄花不同花期;C:雌花不同花期;D:种子不同发育时期及不同部位(1:早期发育种子;2:中期发育种子;3:后期发育种子;4:干种子;5:中期发育的种子子叶;6:中期发育种子胚轴;7:中期发育种子种皮);E:ScHSP20基因在渗透(OS)、高温(HT)、低温(LT)胁迫下的表达
Fig. 5 Expressions of ScHSP20 gene in different tissues and under different stresses A: Leaves and fruits of Simondwood. B: Male flowers at different flowering stages. C: Different flowering stages of female flowers in Simondmu. D: Different stages and parts of seed development in Simondwood(1: Early developmental seeds. 2: Mid stage developmental seeds. 3: Late developmental seeds. 4: Dry seeds. 5: Middle stage development of seed cotyledons. 6: Mid developmental seed hypocotyl. 7: Mid stage development of seed coat). E: The expression of ScHSP20 gene under osmotic(OS), high temperature(HT), and low temperature(LT)stress
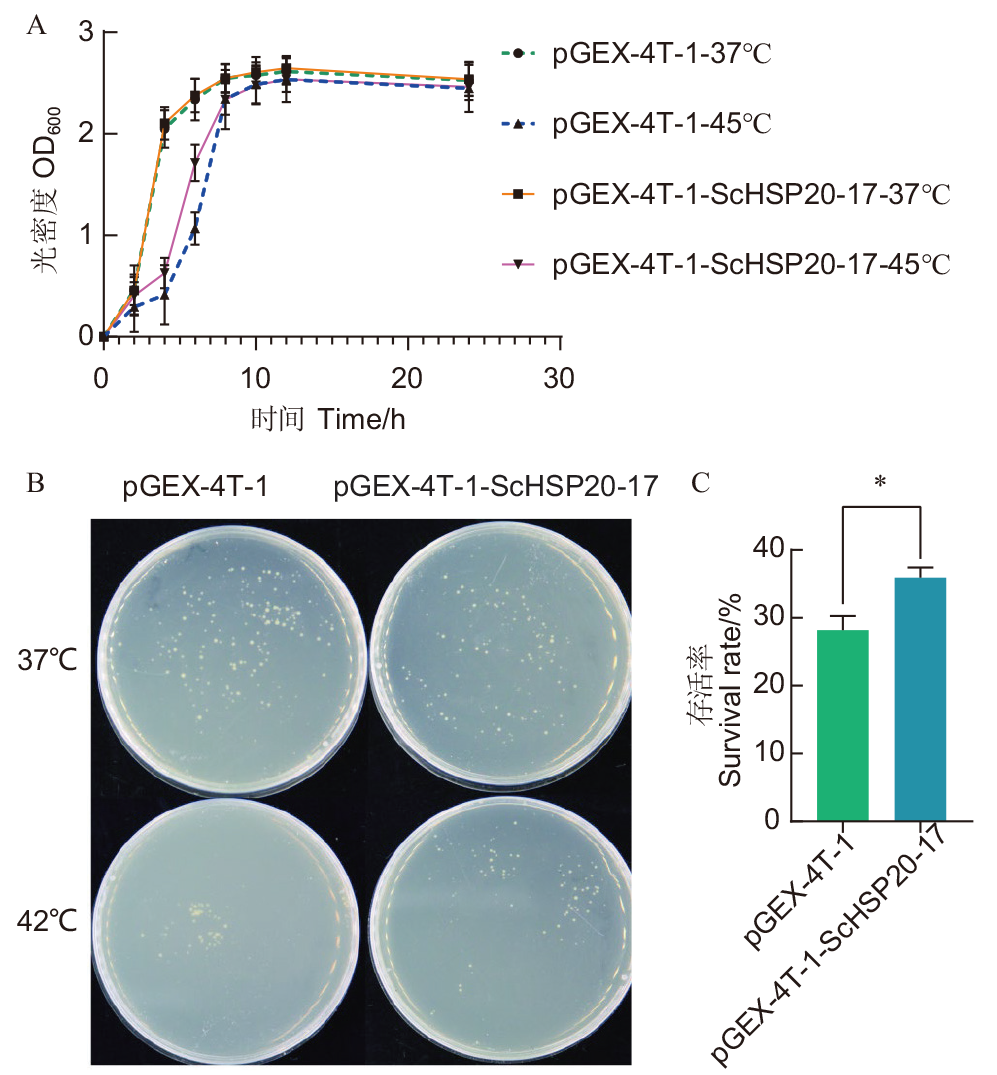
图9 ScHSP20-17异源表达对大肠杆菌高温胁迫耐受性的影响 A:高温胁迫处理下重组菌与对照菌的生长曲线;B:处理前后对照菌与重组菌的生长情况;C:高温胁迫处理前后对照菌与重组菌的存活率统计。*表示在P<0.05水平差异显著
Fig. 9 Effects of heterologous expression of ScHSP20-17 gene on the tolerance of Escherichia coli to high temperature stress A: rowth curves of recombinant bacteria and control bacteria under high temperature stress. B: Growth of control and recombinant bacteria before and after treatment.C: Statistics of survival rate of control bacteria and recombinant bacteria before and after high temperature stress treatment. * indicate significant differences at P<0.05

图11 ScHSP20-17异源表达对烟草高温胁迫耐受性的影响 A:荧光蛋白灯照射下未注射菌液的烟草成像;B:荧光蛋白灯照射下注射菌液的烟草成像;C:野生型烟草(WT)、注射空载(PRI)菌液的烟草和注射携带PRI-ScHSP20-17-GFP载体菌液的烟草叶片RT-qPCR分析;D-E:野生型烟草;F:瞬转空载(PRI)的烟草;G:瞬转ScHSP20-17的烟草;H:MDA含量测定
Fig. 11 Effects of heterologous expression of ScHSP20-17 gene on the tolerance of tobacco to high temperature stress A: Image of tobacco without injection of bacterial liquid under fluorescent protein lamp irradiation. B: Image of tobacco with injection of bacterial liquid under fluorescent protein lamp irradiation. C: Wild type tobacco(WT), tobacco with injection of empty bacterial liquid(PRI), and tobacco leaves with injection of bacterial liquid carrying PRI-ScHSP20-17-GFP vector qRT PCR analysis. D-E: Wild type tobacco. F: Tobacco with instantaneous rotation of empty liquid(PRI). G: Tobacco with instantaneous rotation of ScHSP20-17. H: MDA content determination
| [1] | Zheng LM, Wu WY, Gao YF, et al. Integrated transcriptome, small RNA and degradome analysis provide insights into the transcriptional regulatory networks underlying cold acclimation in jojoba[J]. Sci Hortic, 2022, 299. |
| [2] | Kawiński A, Miklaszewska M, Stelter S, et al. Lipases of germinating jojoba seeds efficiently hydrolyze triacylglycerols and wax esters and display wax ester-synthesizing activity[J]. BMC Plant Biol, 2021, 21(1): 50. |
| [3] | Gad HA, Roberts A, Hamzi SH, et al. Jojoba oil: An updated comprehensive review on chemistry, pharmaceutical uses, and toxicity[J]. Polymers, 2021, 13(11): 1711. |
| [4] | Pazyar N, Yaghoobi R, Ghassemi MR, et al. Jojoba in dermatology: a succinct review[J]. G Ital Dermatol Venereol., 2013, 148(6): 687-691. |
| [5] | Zhu JK. Abiotic stress signaling and responses in plants[J]. Cell, 2016, 167(2): 313-324. |
| [6] |
Neta-Sharir I, Isaacson T, Lurie S, et al. Dual role for tomato heat shock protein 21: protecting photosystem II from oxidative stress and promoting color changes during fruit maturation[J]. Plant Cell, 2005, 17(6): 1829-1838.
doi: 10.1105/tpc.105.031914 pmid: 15879560 |
| [7] | Yu JH, Cheng Y, Feng K, et al. Genome-wide identification and expression profiling of tomato HSP20 gene family in response to biotic and abiotic stresses[J]. Front Plant Sci, 2016, 7: 1215. |
| [8] |
Wang WX, Vinocur B, Shoseyov O, et al. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response[J]. Trends Plant Sci, 2004, 9(5): 244-252.
doi: 10.1016/j.tplants.2004.03.006 pmid: 15130550 |
| [9] |
Waters ER. The evolution, function, structure, and expression of the plant sHSPs[J]. J Exp Bot, 2013, 64(2): 391-403.
doi: 10.1093/jxb/ers355 pmid: 23255280 |
| [10] | Qi XY, Di ZX, Li YY, et al. Genome-wide identification and expression profiling of heat shock protein 20 gene family in Sorbus pohuashanensis(Hance)hedl under abiotic stress[J]. Genes, 2022, 13(12): 2241. |
| [11] |
Jaya N, Garcia V, Vierling E. Substrate binding site flexibility of the small heat shock protein molecular chaperones[J]. Proc Natl Acad Sci USA, 2009, 106(37): 15604-15609.
doi: 10.1073/pnas.0902177106 pmid: 19717454 |
| [12] |
Sun WN, Van Montagu M, Verbruggen N. Small heat shock proteins and stress tolerance in plants[J]. Biochim Biophys Acta, 2002, 1577(1): 1-9.
doi: 10.1016/s0167-4781(02)00417-7 pmid: 12151089 |
| [13] |
Haslbeck M, Vierling E. A first line of stress defense: small heat shock proteins and their function in protein homeostasis[J]. J Mol Biol, 2015, 427(7): 1537-1548.
doi: 10.1016/j.jmb.2015.02.002 pmid: 25681016 |
| [14] | Ji XR, Yu YH, Ni PY, et al. Genome-wide identification of small heat-shock protein(HSP20)gene family in grape and expression profile during berry development[J]. BMC Plant Biol, 2019, 19(1): 433. |
| [15] |
Ouyang YD, Chen JJ, Xie WB, et al. Comprehensive sequence and expression profile analysis of HSP20 gene family in rice[J]. Plant Mol Biol, 2009, 70(3): 341-357.
doi: 10.1007/s11103-009-9477-y pmid: 19277876 |
| [16] |
Lopes-Caitar VS, Darben LM, et al. Genome-wide analysis of the HSP20 gene family in soybean: comprehensive sequence, genomic organization and expression profile analysis under abiotic and biotic stresses[J]. BMC Genomics, 2013, 14: 577.
doi: 10.1186/1471-2164-14-577 pmid: 23985061 |
| [17] |
Guo M, Liu JH, Lu JP, et al. Genome-wide analysis of the CaHsp20 gene family in pepper: comprehensive sequence and expression profile analysis under heat stress[J]. Front Plant Sci, 2015, 6: 806.
doi: 10.3389/fpls.2015.00806 pmid: 26483820 |
| [18] | Scharf KD, Siddique M, Vierling E. The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing alpha-crystallin domains(Acd proteins)[J]. Cell Stress and Chaperones, 2001, 6(3): 225-237. |
| [19] |
Chen CJ, Chen H, Zhang Y, et al. TBtools: An integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [20] | Sturtevant D, Lu SP, Zhou ZW, et al. The genome of jojoba(Simmondsia chinensis): A taxonomically isolated species that directs wax ester accumulation in its seeds[J]. Sci Adv, 2020, 6(11): eaay3240. |
| [21] |
孙小倩, 王佳蕊, 陈庆富, 等. 苦荞转录因子FtMYBF的克隆、亚细胞定位及表达分析[J]. 生物技术通报, 2021, 37(3): 10-17.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-0861 |
| Sun XQ, Wang JR, Chen QF, et al. Cloning, subcellular localization, and expression analysis of the transcription factor FtMYBF from tartary buckwheat[J]. Biotechnol Bulletin, 2021, 37(3): 10-17. | |
| [22] | Yao FW, Song CH, Wang HT, et al. Genome-wide characterization of the HSP20 gene family identifies potential members involved in temperature stress response in apple[J]. Front Genet, 2020, 11: 609184. |
| [23] | Aghdam MS, Sevillano L, Flores FB, et al. Heat shock proteins as biochemical markers for postharvest chilling stress in fruits and vegetables[J]. Sci Hortic, 2013, 160: 54-64. |
| [24] | Hu YP, Zhang TT, Liu Y, et al. Pumpkin(Cucurbita moschata)HSP20 gene family identification and expression under heat stress[J]. Front Genet, 2021, 12: 753953. |
| [25] | Huang JJ, Hai ZX, Wang RY, et al. Genome-wide analysis of HSP20 gene family and expression patterns under heat stress in cucumber(Cucumis sativus L.)[J]. Front Plant Sci, 2022, 13: 968418. |
| [26] | Lian XD, Wang QP, Li TH, et al. Phylogenetic and transcriptional analyses of the HSP20 gene family in peach revealed that PpHSP20-32 is involved in plant height and heat tolerance[J]. Int J Mol Sci, 2022, 23(18): 10849. |
| [27] |
Nagaraju M, Reddy PS, Kumar SA, et al. Genome-wide identification and transcriptional profiling of small heat shock protein gene family under diverse abiotic stress conditions in Sorghum bicolor(L.)[J]. Int J Biol Macromol, 2020, 142: 822-834.
doi: S0141-8130(19)35485-6 pmid: 31622710 |
| [28] |
Xu GX, Guo CC, Shan HY, et al. Divergence of duplicate genes in exon-intron structure[J]. Proc Natl Acad Sci USA, 2012, 109(4): 1187-1192.
doi: 10.1073/pnas.1109047109 pmid: 22232673 |
| [29] |
Mattick JS, Gagen MJ. The evolution of controlled multitasked gene networks: the role of introns and other noncoding RNAs in the development of complex organisms[J]. Mol Biol Evol, 2001, 18(9): 1611-1130.
doi: 10.1093/oxfordjournals.molbev.a003951 pmid: 11504843 |
| [30] | Chung BYW, Simons C, Firth AE, et al. Effect of 5'UTR introns on gene expression in Arabidopsis thaliana[J]. BMC Genomics, 2006, 7: 120. |
| [31] | Sarkar NK, Kim YK, Grover A. Rice shsp genes: genomic organization and expression profiling under stress and development[J]. BMC Genom, 2009, 10(1): 393. |
| [32] |
Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis[J]. Science, 2000, 290(5499): 2114-2117.
doi: 10.1126/science.290.5499.2114 pmid: 11118139 |
| [33] |
Conant GC, Wolfe KH. Turning a hobby into a job: how duplicated genes find new functions[J]. Nat Rev Genet, 2008, 9(12): 938-950.
doi: 10.1038/nrg2482 pmid: 19015656 |
| [34] | De Grassi A, Lanave C, Saccone C. Genome duplication and gene-family evolution: the case of three OXPHOS gene families[J]. Gene, 2008, 421(1/2): 1-6. |
| [35] | Flagel LE, Wendel JF. Gene duplication and evolutionary novelty in plants[J]. The New Phytol, 2009, 183(3): 557-564. |
| [36] | Hua YG, Liu Q, Zhai YF, et al. Genome-wide analysis of the HSP20 gene family and its response to heat and drought stress in Coix(Coix lacryma-jobi L.)[J]. BMC Genomics, 2023, 24(1): 478. |
| [37] |
Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters[J]. Trends Plant Sci, 2005, 10(2): 88-94.
doi: 10.1016/j.tplants.2004.12.012 pmid: 15708346 |
| [38] | Tian C, Zhang ZY, Huang Y, et al. Functional characterization of the Pinellia ternata cytoplasmic class II small heat shock protein gene PtsHSP17.2 via promoter analysis and overexpression in tobacco[J]. Plant Physiol Biochem, 2022, 177: 1-9. |
| [1] | 王玉书, 赵琳琳, 赵爽, 胡琦, 白慧霞, 王欢, 曹业萍, 范震宇. 大白菜BrCYP83B1基因的克隆及表达分析[J]. 生物技术通报, 2024, 40(6): 152-160. |
| [2] | 闫欢欢, 尚怡彤, 王丽红, 田学琴, 廖海艳, 曾斌, 胡志宏. 米曲霉异源表达合成虫草素[J]. 生物技术通报, 2024, 40(6): 290-298. |
| [3] | 常雪瑞, 王田田, 王静. 辣椒E2基因家族的鉴定及分析[J]. 生物技术通报, 2024, 40(6): 238-250. |
| [4] | 刘蓉, 田闵玉, 李光泽, 谭成方, 阮颖, 刘春林. 甘蓝型油菜REVEILLE家族鉴定及诱导表达分析[J]. 生物技术通报, 2024, 40(6): 161-171. |
| [5] | 吴泽航, 杨中义, 鄢毅铖, 贾永红, 吴月燕, 谢晓鸿. 比利时杜鹃花类黄酮3'-羟化酶(F3'H)基因克隆及功能分析[J]. 生物技术通报, 2024, 40(6): 251-259. |
| [6] | 郝思怡, 张君珂, 王斌, 曲朋燕, 李瑞得, 程春振. 香蕉ELF3的克隆与表达分析[J]. 生物技术通报, 2024, 40(5): 131-140. |
| [7] | 李嘉欣, 李鸿燕, 刘丽娥, 张恬, 周武. 沙棘NRAMP基因家族鉴定及铅胁迫下表达分析[J]. 生物技术通报, 2024, 40(5): 191-202. |
| [8] | 潘萍萍, 徐志浩, 张怡雯, 李青, 王忠华. 多花黄精查尔酮合酶PcCHS的原核表达、亚细胞定位及表达分析[J]. 生物技术通报, 2024, 40(5): 280-289. |
| [9] | 张震, 李清, 徐菁, 陈凯园, 张春芝, 祝光涛. 马铃薯线粒体靶向表达载体的构建与应用[J]. 生物技术通报, 2024, 40(5): 66-73. |
| [10] | 娄银, 高浩竣, 王茜, 牛景萍, 王敏, 杜维俊, 岳爱琴. 大豆GmHMGS基因的鉴定及表达模式分析[J]. 生物技术通报, 2024, 40(4): 110-121. |
| [11] | 陈春林, 李白雪, 李金玲, 杜清洁, 李猛, 肖怀娟. 甜瓜CmEPF基因家族的鉴定及表达分析[J]. 生物技术通报, 2024, 40(4): 130-138. |
| [12] | 肖雅茹, 贾婷婷, 罗丹, 武喆, 李丽霞. 黄瓜CsERF025L转录因子的克隆及表达分析[J]. 生物技术通报, 2024, 40(4): 159-166. |
| [13] | 杨伟成, 孙岩, 杨倩, 王壮琳, 马菊花, 薛金爱, 李润植. 陆地棉FAX家族的全基因组鉴定及GhFAX1的功能分析[J]. 生物技术通报, 2024, 40(3): 155-169. |
| [14] | 杨艳, 胡洋, 刘霓如, 殷璐, 杨锐, 王鹏飞, 穆霄鹏, 张帅, 程春振, 张建成. ‘红满堂’苹果MbbZIP43基因的克隆与功能研究[J]. 生物技术通报, 2024, 40(2): 146-159. |
| [15] | 任延靖, 张鲁刚, 赵孟良, 李江, 邵登魁. 白菜种子cDNA酵母文库的构建及BrTTG1互作蛋白的筛选及分析[J]. 生物技术通报, 2024, 40(2): 223-232. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||