








生物技术通报 ›› 2024, Vol. 40 ›› Issue (6): 23-33.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1164
王迪1,2( ), 张晓宇1,2, 宋宇鑫1,2, 郑东然1,2, 田静1,2, 李玉花1,2, 王宇1,2, 吴昊1,2(
), 张晓宇1,2, 宋宇鑫1,2, 郑东然1,2, 田静1,2, 李玉花1,2, 王宇1,2, 吴昊1,2( )
)
收稿日期:2023-12-10
出版日期:2024-06-26
发布日期:2024-06-24
通讯作者:
吴昊,男,博士,讲师,研究方向:药用植物生物反应器应用;E-mail: wuhao@nefu.edu.cn作者简介:王迪,女,硕士研究生,研究方向:植物细胞全能性转录因子;E-mail: 976332140@qq.com
基金资助:
WANG Di ZHANG Xiao-yu SONG Yu-xin ZHENG Dong-ran TIAN Jing LI Yu-hua WANG Yu WU Hao1,2( )
)
Received:2023-12-10
Published:2024-06-26
Online:2024-06-24
摘要:
植物细胞在适宜的培养条件下展现出发育成完整新个体的潜能,这种潜能被称为植物细胞全能性。基于细胞全能性的组织培养技术,在植物无性繁殖和遗传改良领域得到了广泛应用。非生物胁迫、植物激素与转录因子协同调控植物离体再生过程。其中,在植物再生过程中起主导作用的转录因子,被称为细胞全能性转录因子。近年来,关于细胞全能性转录因子调控植物离体再生的分子机制研究已取得显著进展。众多转录因子被鉴定出来,并在提高植物遗传转化效率的研究中得到了初步应用。本文按照植物细胞全能性转录因子在植物再生中的功能进行分类梳理,综述近几年植物离体再生过程中信号转导机制,总结植物细胞全能性转录因子在提高植物遗传转化效率方面的作用,并对其应用前景进行展望,为建立有效的无性繁殖体系提供科学依据。
王迪, 张晓宇, 宋宇鑫, 郑东然, 田静, 李玉花, 王宇, 吴昊. 细胞全能性转录因子调控植物组培再生的分子机制研究进展[J]. 生物技术通报, 2024, 40(6): 23-33.
WANG Di ZHANG Xiao-yu SONG Yu-xin ZHENG Dong-ran TIAN Jing LI Yu-hua WANG Yu WU Hao. Advances in the Molecular Mechanisms of Plant Tissue Culture and Regeneration Regulated by Totipotency-related Transcription Factors[J]. Biotechnology Bulletin, 2024, 40(6): 23-33.
| 转录因子名称Name of transcription factor | 结构域示意图Schematic diagram of domain | 三级结构预测图Tertiary structure prediction diagram |
|---|---|---|
| Homeobox |  | 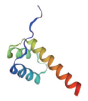 |
| AP2/ERF |  | 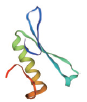 |
| LOB |  | 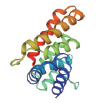 |
| LEC |  | 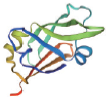 |
表1 主要转录因子结构域及三级结构预测图
Table 1 Tertiary structure prediction diagram of major transcription factor domains
| 转录因子名称Name of transcription factor | 结构域示意图Schematic diagram of domain | 三级结构预测图Tertiary structure prediction diagram |
|---|---|---|
| Homeobox |  |  |
| AP2/ERF |  |  |
| LOB |  |  |
| LEC |  |  |
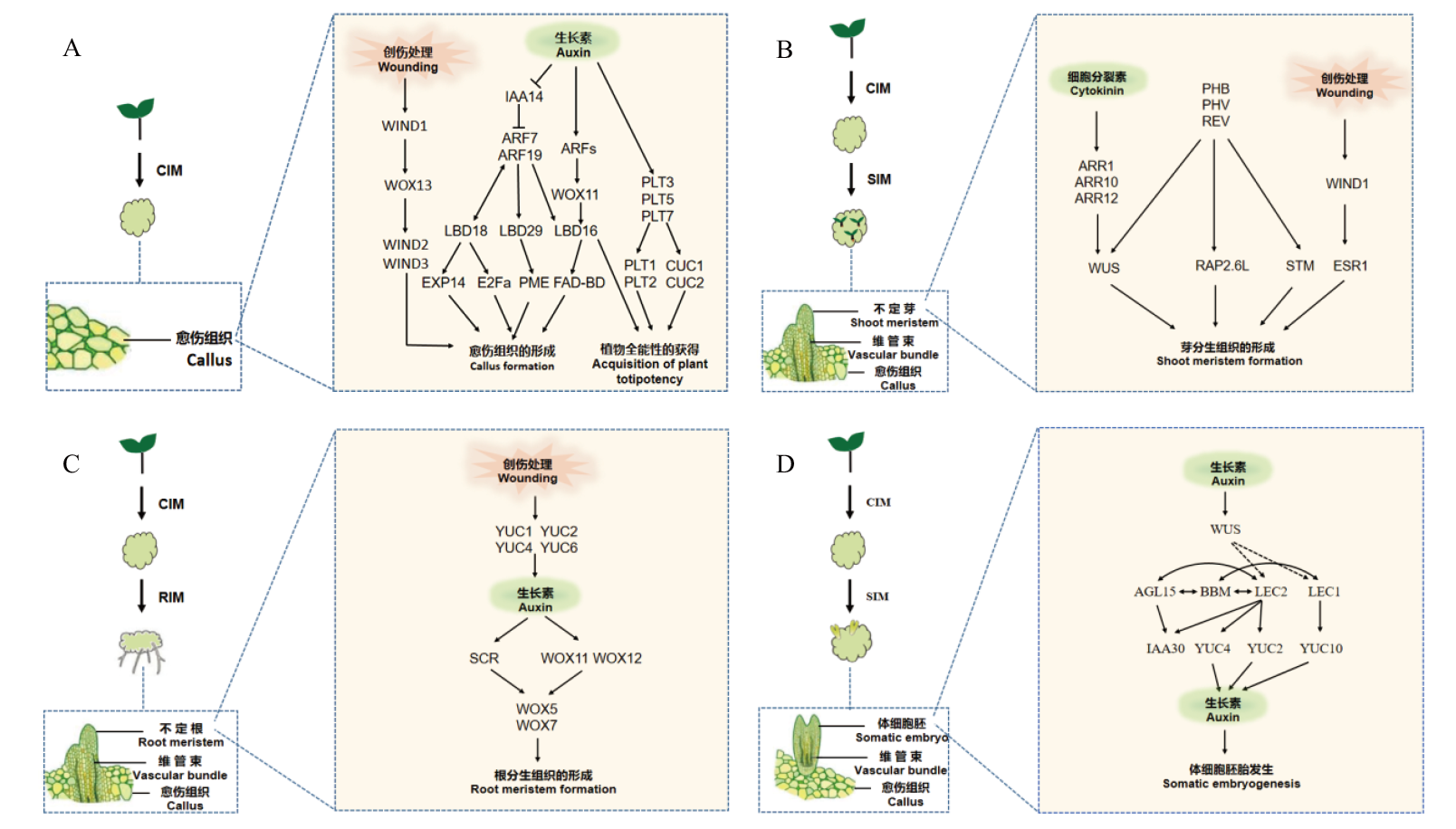
图2 植物组培再生过程中相关转录因子的调控机制 A:愈伤组织形成过程中相关转录因子调控机制;B:不定芽再生过程中相关转录因子调控机制;C:根分生组织形成过程中相关转录因子调控机制;D:体细胞胚胎发生过程中相关转录因子调控机制
Fig. 2 Regulation mechanism of related transcription factors in plant tissue culture regeneration A: Regulation mechanism of related transcription factors during callus formation; B: regulation mechanism of related transcription factors in shoot regeneration; C: regulation mechanism of related transcription factors during root meristem formation; D: regulation mechanism of related transcription factors in somatic embryogenesis
| [1] | 胡彦, 赵艳. 植物组织培养技术的应用以及在培养过程中存在的问题[J]. 陕西师范大学学报: 自然科学版, 2004, 32(S1): 130-134. |
| Hu Y, Zhao Y. Application of plant tissue culture technology and problems existing in the process of culture[J]. J Shaanxi Norm Univ Nat Sci Ed, 2004, 32(S1): 130-134. | |
| [2] | 许智宏, 张宪省, 苏英华, 等. 植物细胞全能性和再生[J]. 中国科学: 生命科学, 2019, 49(10): 1282-1300. |
| Xu ZH, Zhang XS, Su YH, et al. Plant cell totipotency and regeneration[J]. Sci Sin Vitae, 2019, 49(10): 1282-1300. | |
| [3] |
Ikeuchi M, Favero DS, Sakamoto Y, et al. Molecular mechanisms of plant regeneration[J]. Annu Rev Plant Biol, 2019, 70: 377-406.
doi: 10.1146/annurev-arplant-050718-100434 pmid: 30786238 |
| [4] |
Birnbaum KD, Alvarado AS. Slicing across Kingdoms: regeneration in plants and animals[J]. Cell, 2008, 132(4): 697-710.
doi: 10.1016/j.cell.2008.01.040 pmid: 18295584 |
| [5] |
Méndez-Hernández HA, Ledezma-Rodríguez M, Avilez-Montalvo RN, et al. Signaling overview of plant somatic embryogenesis[J]. Front Plant Sci, 2019, 10: 77.
doi: 10.3389/fpls.2019.00077 pmid: 30792725 |
| [6] |
Su YH, Tang LP, Zhao XY, et al. Plant cell totipotency: insights into cellular reprogramming[J]. J Integr Plant Biol, 2021, 63(1): 228-243.
doi: 10.1111/jipb.12972 |
| [7] |
Xu J, Hofhuis H, Heidstra R, et al. A molecular framework for plant regeneration[J]. Science, 2006, 311(5759): 385-388.
pmid: 16424342 |
| [8] | Junker A, Mönke G, Rutten T, et al. Elongation-related functions of LEAFY COTYLEDON1 during the development of Arabidopsis thaliana[J]. Plant J, 2012, 71(3): 427-442. |
| [9] |
Braybrook SA, Stone SL, Park S, et al. Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis[J]. Proc Natl Acad Sci USA, 2006, 103(9): 3468-3473.
doi: 10.1073/pnas.0511331103 pmid: 16492731 |
| [10] |
Lotan T, Ohto M, Yee KM, et al. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells[J]. Cell, 1998, 93(7): 1195-1205.
doi: 10.1016/s0092-8674(00)81463-4 pmid: 9657152 |
| [11] | Ren C, Zhang Z, Wang Y, et al. Genome-wide identification and characterization of the NF-Y gene family in grape(Vitis vinifera L.)[J]. BMC Genomics, 2016, 17(1): 605. |
| [12] |
Horstman A, Li MF, Heidmann I, et al. The BABY BOOM transcription factor activates the LEC1-ABI3-FUS3-LEC2 network to induce somatic embryogenesis[J]. Plant Physiol, 2017, 175(2): 848-857.
doi: 10.1104/pp.17.00232 pmid: 28830937 |
| [13] | Ledwoń A, Gaj MD. LEAFY COTYLEDON2 gene expression and auxin treatment in relation to embryogenic capacity of Arabidopsis somatic cells[J]. Plant Cell Rep, 2009, 28(11): 1677-1688. |
| [14] | Li K, Wang J, Liu CL, et al. Expression of AtLEC2 and AtIPTs promotes embryogenic callus formation and shoot regeneration in tobacco[J]. BMC Plant Biol, 2019, 19(1): 314. |
| [15] | Gallois JL, Nora FR, Mizukami Y, et al. WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem[J]. Genes Dev, 2004, 18(4): 375-380. |
| [16] | Nair S, Bahn JH, Lee G, et al. A homeobox transcription factor scarecrow(SCRO)negatively regulates pdf neuropeptide expression through binding an identified cis-acting element in Drosophila melanogaster[J]. Mol Neurobiol, 2020, 57(4): 2115-2130. |
| [17] |
Hu XM, Xu L. Transcription factors WOX11/12 directly activate WOX5/7 to promote root primordia initiation and organogenesis[J]. Plant Physiol, 2016, 172(4): 2363-2373.
pmid: 27784768 |
| [18] | Liu JC, Sheng LH, Xu YQ, et al. WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis[J]. Plant Cell, 2014, 26(3): 1081-1093. |
| [19] | Meng WJ, Cheng ZJ, Sang YL, et al. Type-B ARABIDOPSIS RESPONSE REGULATORs specify the shoot stem cell niche by dual regulation of WUSCHEL[J]. Plant Cell, 2017, 29(6): 1357-1372. |
| [20] | Su YH, Zhao XY, Liu YB, et al. Auxin-induced WUS expression is essential for embryonic stem cell renewal during somatic embryogenesis in Arabidopsis[J]. Plant J, 2009, 59(3): 448-460. |
| [21] | Zhang TQ, Lian H, Zhou CM, et al. A two-step model for de novo activation of WUSCHEL during plant shoot regeneration[J]. Plant Cell, 2017, 29(5): 1073-1087. |
| [22] | Kim JY, Yang W, Forner J, et al. Epigenetic reprogramming by histone acetyltransferase HAG1/AtGCN5 is required for pluripotency acquisition in Arabidopsis[J]. EMBO J, 2018, 37(20): e98726. |
| [23] |
Zhang X, Zong J, Liu JH, et al. Genome-wide analysis of WOX gene family in rice, sorghum, maize, Arabidopsis and poplar[J]. J Integr Plant Biol, 2010, 52(11): 1016-1026.
doi: 10.1111/j.1744-7909.2010.00982.x |
| [24] | Luo HM, Xu ZC, Song JY, et al. Research and perspectives on AP2/ERF transcription factors in medicinal plants[J]. Chin Sci Bull, 2015, 60(14): 1272-1284. |
| [25] | Campbell SSB. Studies of the role of Baby Boom(BBM)in embryogenesis in Arabidopsis thaliana and Brassica napus[D]. University of Guelph(Canada). 2009. |
| [26] | Jha P, Kumar V. BABY BOOM(BBM): a candidate transcription factor gene in plant biotechnology[J]. Biotechnol Lett, 2018, 40(11/12): 1467-1475. |
| [27] | Li MF, Wrobel-Marek J, Heidmann I, et al. Auxin biosynthesis maintains embryo identity and growth during BABY BOOM-induced somatic embryogenesis[J]. Plant Physiol, 2022, 188(2): 1095-1110. |
| [28] | Srinivasan C, Liu ZR, Heidmann I, et al. Heterologous expression of the baby boom ap2/erf transcription factor enhances the regeneration capacity of tobacco(Nicotiana tabacum L.)[J]. Planta, 2007, 225(2): 341-351. |
| [29] | Zhou RN, Zhao YJ, Cheng P, et al. GmBBM7 promotes callus and root growth during somatic embryogenesis of soybean(Glycine max)[J]. Biotechnol Biotechnol Equip, 2023, 37(1). |
| [30] | Iwase A, Harashima H, Ikeuchi M, et al. WIND1 promotes shoot regeneration through transcriptional activation of ENHANCER OF SHOOT REGENERATION1 in Arabidopsis[J]. Plant Cell, 2017, 29(1): 54-69. |
| [31] | Iwase A, Mita K, Nonaka S, et al. WIND1-based acquisition of regeneration competency in Arabidopsis and rapeseed[J]. J Plant Res, 2015, 128(3): 389-397. |
| [32] | Iwase A, Mitsuda N, Koyama T, et al. The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis[J]. Curr Biol, 2011, 21(6): 508-514. |
| [33] | Iwase A, Mitsuda N, Ikeuchi M, et al. Arabidopsis WIND1 induces callus formation in rapeseed, tomato, and tobacco[J]. Plant Signal Behav, 2013, 8(12): e27432. |
| [34] |
Kareem A, Durgaprasad K, Sugimoto K, et al. PLETHORA genes control regeneration by a two-step mechanism[J]. Curr Biol, 2015, 25(8): 1017-1030.
doi: 10.1016/j.cub.2015.02.022 pmid: 25819565 |
| [35] |
Bustillo-Avendaño E, Ibáñez S, Sanz O, et al. Regulation of hormonal control, cell reprogramming, and patterning during de novo root organogenesis[J]. Plant Physiol, 2018, 176(2): 1709-1727.
doi: 10.1104/pp.17.00980 pmid: 29233938 |
| [36] |
Ikeuchi M, Iwase A, Rymen B, et al. Wounding triggers callus formation via dynamic hormonal and transcriptional changes[J]. Plant Physiol, 2017, 175(3): 1158-1174.
doi: 10.1104/pp.17.01035 pmid: 28904073 |
| [37] |
Tsuwamoto R, Yokoi S, Takahata Y. Arabidopsis EMBRYOMAKER encoding an AP2 domain transcription factor plays a key role in developmental change from vegetative to embryonic phase[J]. Plant Mol Biol, 2010, 73(4-5): 481-492.
doi: 10.1007/s11103-010-9634-3 pmid: 20405311 |
| [38] | Lee K, Park OS, Seo PJ. Arabidopsis ATXR2 deposits H3K36me3 at the promoters of LBD genes to facilitate cellular dedifferentiation[J]. Sci Signal, 2017, 10(507): eaan0316. |
| [39] | Xu CY, Cao HF, Zhang QQ, et al. Control of auxin-induced callus formation by bZIP59-LBD complex in Arabidopsis regeneration[J]. Nat Plants, 2018, 4(2): 108-115. |
| [40] | Xu CY, Cao HF, Xu EJ, et al. Genome-wide identification of Arabidopsis LBD29 target genes reveals the molecular events behind auxin-induced cell reprogramming during callus formation[J]. Plant Cell Physiol, 2018, 59(4): 744-755. |
| [41] |
Okushima Y, Fukaki H, Onoda M, et al. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis[J]. Plant Cell, 2007, 19(1): 118-130.
doi: 10.1105/tpc.106.047761 pmid: 17259263 |
| [42] | Atta R, Laurens L, Boucheron-Dubuisson E, et al. Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro[J]. Plant J, 2009, 57(4): 626-644. |
| [43] | Van Tuong Huan L, Takamura T, Tanaka M. Callus formation and plant regeneration from callus through somatic embryo structures in Cymbidium orchid[J]. Plant Sci, 2004, 166(6): 1443-1449. |
| [44] | Heyman J, Cools T, Canher B, et al. The heterodimeric transcription factor complex ERF115-PAT1 grants regeneration competence[J]. Nat Plants, 2016, 2(11): 16165. |
| [45] | Liu J, Hu XM, Qin P, et al. The WOX11-LBD16 pathway promotes pluripotency acquisition in callus cells during de novo shoot regeneration in tissue culture[J]. Plant Cell Physiol, 2018, 59(4): 734-743. |
| [46] | Fukaki H, Nakao Y, Okushima Y, et al. Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis[J]. Plant J, 2005, 44(3): 382-395. |
| [47] | Fan MZ, Xu CY, Xu K, et al. Lateral organ boundaries domain transcription factors direct callus formation in Arabidopsis regeneration[J]. Cell Res, 2012, 22(7): 1169-1180. |
| [48] | Pandey SK, Lee HW, Kim MJ, et al. LBD18 uses a dual mode of a positive feedback loop to regulate ARF expression and transcriptional activity in Arabidopsis[J]. Plant J, 2018, 95(2): 233-251. |
| [49] | Berckmans B, Vassileva V, Schmid SPC, et al. Auxin-dependent cell cycle reactivation through transcriptional regulation of Arabidopsis E2Fa by lateral organ boundary proteins[J]. Plant Cell, 2011, 23(10): 3671-3683. |
| [50] | Lee HW, Kim MJ, Kim NY, et al. LBD18 acts as a transcriptional activator that directly binds to the EXPANSIN14 promoter in promoting lateral root emergence of Arabidopsis[J]. Plant J, 2013, 73(2): 212-224. |
| [51] |
Gordon SP, Heisler MG, Reddy GV, et al. Pattern formation during de novo assembly of the Arabidopsis shoot meristem[J]. Development, 2007, 134(19): 3539-3548.
doi: 10.1242/dev.010298 pmid: 17827180 |
| [52] | Higuchi M, Pischke MS, Mähönen AP, et al. In planta functions of the Arabidopsis cytokinin receptor family[J]. Proc Natl Acad Sci USA, 2004, 101(23): 8821-8826. |
| [53] | Shi BH, Zhang C, Tian CH, et al. Two-step regulation of a meristematic cell population acting in shoot branching in Arabidopsis[J]. PLoS Genet, 2016, 12(7): e1006168. |
| [54] | Che P, Lall S, Nettleton D, et al. Gene expression programs during shoot, root, and callus development in Arabidopsis tissue culture[J]. Plant Physiol, 2006, 141(2): 620-637. |
| [55] |
Yang SQ, Poretska O, Sieberer T. Altered meristem program1 restricts shoot meristem proliferation and regeneration by limiting hd-zip iii-mediated expression of rap2.6l[J]. Plant Physiol, 2018, 177(4): 1580-1594.
doi: 10.1104/pp.18.00252 pmid: 29884678 |
| [56] | Chen LQ, Tong JH, Xiao LT, et al. YUCCA-mediated auxin biogenesis is required for cell fate transition occurring during de novo root organogenesis in Arabidopsis[J]. J Exp Bot, 2016, 67(14): 4273-4284. |
| [57] | Sheng LH, Hu XM, Du YJ, et al. Non-canonical WOX11-mediated root branching contributes to plasticity in Arabidopsis root system architecture[J]. Development, 2017, 144(17): 3126-3133. |
| [58] |
Ulmasov T, Hagen G, Guilfoyle TJ. Activation and repression of transcription by auxin-response factors[J]. Proc Natl Acad Sci USA, 1999, 96(10): 5844-5849.
pmid: 10318972 |
| [59] |
Forzani C, Aichinger E, Sornay E, et al. WOX5 suppresses CYCLIN D activity to establish quiescence at the center of the root stem cell niche[J]. Curr Biol, 2014, 24(16):1939-1944.
doi: 10.1016/j.cub.2014.07.019 pmid: 25127220 |
| [60] |
Fehér A. Somatic embryogenesis - Stress-induced remodeling of plant cell fate[J]. Biochim Biophys Acta, 2015, 1849(4): 385-402.
doi: 10.1016/j.bbagrm.2014.07.005 pmid: 25038583 |
| [61] |
Harding EW, Tang WN, Nichols KW, et al. Expression and maintenance of embryogenic potential is enhanced through constitutive expression of AGAMOUS-Like 15[J]. Plant Physiol, 2003, 133(2): 653-663.
doi: 10.1104/pp.103.023499 pmid: 14512519 |
| [62] |
Luo GB, Palmgren M. GRF-GIF chimeras boost plant regeneration[J]. Trends Plant Sci, 2021, 26(3): 201-204.
doi: 10.1016/j.tplants.2020.12.001 pmid: 33349565 |
| [63] |
Debernardi JM, Tricoli DM, Ercoli MF, et al. A GRF-GIF chimeric protein improves the regeneration efficiency of transgenic plants[J]. Nat Biotechnol, 2020, 38(11): 1274-1279.
doi: 10.1038/s41587-020-0703-0 pmid: 33046875 |
| [64] |
Liu XM, Bie XM, Lin XL, et al. Uncovering the transcriptional regulatory network involved in boosting wheat regeneration and transformation[J]. Nat Plants, 2023, 9(6): 908-925.
doi: 10.1038/s41477-023-01406-z pmid: 37142750 |
| [65] |
Wang K, Shi L, Liang XN, et al. The gene TaWOX5 overcomes genotype dependency in wheat genetic transformation[J]. Nat Plants, 2022, 8(2): 110-117.
doi: 10.1038/s41477-021-01085-8 pmid: 35027699 |
| [66] |
Krakowsky MD, Lee M, Garay L, et al. Quantitative trait loci for callus initiation and totipotency in maize(Zea mays L.)[J]. Theor Appl Genet, 2006, 113(5): 821-830.
pmid: 16896717 |
| [67] | Kausch AP, Adams TR, Mangano M, et al. Effects of microprojectile bombardment on embryogenic suspension cell cultures of maize(Zea mays L.) used for genetic transformation[J]. Planta, 1995, 196(3): 501-509. |
| [68] |
McFarland FL, Collier R, Walter N, et al. A key to totipotency: Wuschel-like homeobox 2a unlocks embryogenic culture response in maize(Zea mays L.)[J]. Plant Biotechnol J, 2023, 21(9): 1860-1872.
doi: 10.1111/pbi.14098 pmid: 37357571 |
| [69] | Muppala S, Gudlavalleti PK, Pagidoju S, et al. Distinctive response of maize(Zea mays L.) genotypes in vitro with the acceleration of phytohormones[J]. J Plant Biotechnol, 2020, 47(1): 26-39. |
| [70] | Nelson-Vasilchik K, Hague JP, Tilelli M, et al. Rapid transformation and plant regeneration of sorghum(Sorghum bicolor L.) mediated by altruistic Baby boom and Wuschel2[J]. Vitro Cell Dev Biol Plant, 2022, 58(3): 331-342. |
| [71] | Aregawi K, Shen JQ, Pierroz G, et al. Morphogene-assisted transformation of Sorghum bicolor allows more efficient genome editing[J]. Plant Biotechnol J, 2022, 20(4): 748-760. |
| [72] | Aregawi K, Shen JQ, Pierroz G, et al. Increased engineering and editing efficiency of Sorghum bicolor using morphogene-assisted transformation[J]. In Vitro Cellular and Development Biology-Plant: Journal of the Tissue Culture Association, 2022, 58(4):682-683. |
| [73] | Mao JP, Niu CD, Li K, et al. Melatonin promotes adventitious root formation in apple by promoting the function of MdWOX11[J]. BMC Plant Biol, 2020, 20(1): 536. |
| [74] | Xu XZ, Che QQ, Cheng CX, et al. Genome-wide identification of WOX gene family in apple and a functional analysis of MdWOX4b during adventitious root formation[J]. J Integr Agric, 2022, 21(5): 1332-1345. |
| [75] | 肖旭. 苹果Baby Boom基因在遗传转化和体胚发生中的功能鉴定[D]. 泰安: 山东农业大学, 2023. |
| Xiao X. Functional identification of apple Baby Boom in genetic transformation and somatic embryogenesis[D]. Tai'an: Shandong Agricultural University, 2023. |
| [1] | 花子晴, 周静远, 董合忠. 双子叶植物下胚轴和顶端弯钩发育及其对出苗的调控机制[J]. 生物技术通报, 2024, 40(4): 23-32. |
| [2] | 林鑫焱, 张传忠, 戴兵, 王馨珩, 刘剑锋, 温丽, 徐兴健, 方军. 水稻穗发芽遗传与分子机制的研究进展[J]. 生物技术通报, 2024, 40(1): 24-31. |
| [3] | 位欣欣, 兰海燕. 植物MYB转录因子调控次生代谢及逆境响应的研究进展[J]. 生物技术通报, 2022, 38(8): 12-23. |
| [4] | 张婵, 吴友根, 于靖, 杨东梅, 姚广龙, 杨华庚, 张军锋, 陈萍. 光与茉莉酸信号介导的萜类化合物合成分子机制[J]. 生物技术通报, 2022, 38(8): 32-40. |
| [5] | 孙曼銮, 葛赛, 卜佳, 朱壮彦. 大肠杆菌核糖核酸酶调控机制研究[J]. 生物技术通报, 2022, 38(3): 234-245. |
| [6] | 宗梅, 韩硕, 郭宁, 段蒙蒙, 刘凡, 王桂香. 利用真空渗透和CRISPR/Cas9系统获得非转基因菜薹突变体[J]. 生物技术通报, 2022, 38(10): 159-163. |
| [7] | 张凤, 陈伟. 代谢组学在植物逆境生物学中的研究进展[J]. 生物技术通报, 2021, 37(8): 1-11. |
| [8] | 张婵, 姚广龙, 张军锋, 于靖, 杨东梅, 陈萍, 吴友根. 广藿香百秋李醇分子调控及合成生物学研究进展[J]. 生物技术通报, 2021, 37(8): 55-64. |
| [9] | 钱虹萍, 陈博, 林金星, 崔亚宁. RNA聚合酶II动态调控及其成像技术的研究进展[J]. 生物技术通报, 2021, 37(4): 293-302. |
| [10] | 邹坤, 路丽丽, Collins Asiamah Amponsah, 薛缘, 张少伟, 苏瑛, 赵志辉. 家禽卵泡闭锁机制的研究进展[J]. 生物技术通报, 2020, 36(4): 185-191. |
| [11] | 李泽卿, 刘彩贤, 邢文, 文亚峰. miRNA在植物响应高温胁迫中的研究进展[J]. 生物技术通报, 2020, 36(2): 149-157. |
| [12] | 王小河, 辜夕容, 李杰, 崔瑶. 榧树属植物的无性繁殖技术研究进展[J]. 生物技术通报, 2020, 36(12): 178-187. |
| [13] | 郑文清, 张倩, 杜亮. 短串联靶标模拟技术及其在植物miRNA功能研究中的应用[J]. 生物技术通报, 2020, 36(12): 256-264. |
| [14] | 位明明, 曾霞, 安泽伟, 胡彦师, 黄肖, 李维国. C类花器官特征基因AGAMOUS(AG)调控植物花分生组织维持与终止研究进展[J]. 生物技术通报, 2020, 36(1): 135-143. |
| [15] | 常永芳, 包鹏甲, 褚敏, 吴晓云, 梁春年, 阎萍. LncRNA在哺乳动物毛囊发育调控中的研究进展[J]. 生物技术通报, 2019, 35(8): 205-212. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||