








生物技术通报 ›› 2024, Vol. 40 ›› Issue (7): 150-162.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1090
张明亚( ), 庞胜群(
), 庞胜群( ), 刘玉东, 苏永峰, 牛博文, 韩琼琼
), 刘玉东, 苏永峰, 牛博文, 韩琼琼
收稿日期:2023-11-20
出版日期:2024-07-26
发布日期:2024-07-30
通讯作者:
庞胜群,女,硕士,副教授,硕士生导师,研究方向:蔬菜遗传育种;E-mail: pangshqok@shzu.edu.cn作者简介:张明亚,女,硕士研究生,研究方向:蔬菜遗传育种;E-mail: 2419538996@qq.com
基金资助:
ZHANG Ming-ya( ), PANG Sheng-qun(
), PANG Sheng-qun( ), LIU Yu-dong, SU Yong-feng, NIU Bo-wen, HAN Qiong-qiong
), LIU Yu-dong, SU Yong-feng, NIU Bo-wen, HAN Qiong-qiong
Received:2023-11-20
Published:2024-07-26
Online:2024-07-30
摘要:
【目的】 脂肪酸脱氢酶(fatty acid desaturase,FAD)是一类催化植物不饱和脂肪酸合成的关键酶,在植物生长发育及逆境胁迫响应中起重要作用。鉴定番茄SlFAD基因家族,为番茄SlFAD基因家族功能研究及遗传改良提供理论依据。【方法】 采用生物信息学方法,对其基因家族成员进行鉴定,并对其理化性质、基因结构、系统发育树和表达模式等方面进行研究。【结果】 在番茄基因组中共鉴定出26个SlFAD基因,可分为4个亚族;理化性质分析显示SlFAD蛋白的氨基酸个数在119-912个之间,分子量介于13 288.27-102 522.25 Da,SlFAD蛋白在番茄中主要以碱性蛋白的性质存在,大部分SlFAD家族成员为稳定蛋白和亲水性蛋白;亚细胞定位预测SlFAD基因家族成员主要存在于内质网;基因特征分析显示同一亚族间的成员具有较为相似的基因结构和保守基序;启动子顺式作用元件分析显示数量最多的是光响应与激素响应相关的元件;染色体定位显示26个SlFAD家族成员共分布在10条染色体上,6号和12号染色体上成员最多;二级结构预测显示26个SlFADs家族成员均以α-螺旋和β-转角为主;转录组数据及RT-qPCR分析表明SlFAD1、SlFAD4和SlFAD18基因在成熟花药中高度表达,SlFAD23在根尖中高度表达,说明SlFAD1、SlFAD4和SlFAD18可能参与花药发育,SlFAD23可能参与根尖发育;低温胁迫下,SlFAD6和SlFAD21表达量被抑制,说明SlFAD6和SlFAD21可能与低温响应有关系,SlFAD1和SlFAD14在低温胁迫初期显著上调,随后又被抑制,说明该基因在低温胁迫初期发挥重要作用。【结论】 SlFAD基因家族在番茄根尖、叶片、花药的生长发育过程及低温胁迫中发挥重要作用。
张明亚, 庞胜群, 刘玉东, 苏永峰, 牛博文, 韩琼琼. 番茄FAD基因家族的鉴定与表达分析[J]. 生物技术通报, 2024, 40(7): 150-162.
ZHANG Ming-ya, PANG Sheng-qun, LIU Yu-dong, SU Yong-feng, NIU Bo-wen, HAN Qiong-qiong. Identification and Expression Analysis of FAD Gene Family in Solanum lycopersicum[J]. Biotechnology Bulletin, 2024, 40(7): 150-162.
| 基因名称Gene name | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| Actin | ATCTTGGCTTCCCTCAGCAC | GCATCTCTGGTCCAGTAGGAAAT |
| SlFAD1 | TCTACGGTGTGCCCCTTCTA | AGTGATGGGTGAGTGTGCTG |
| SlFAD4 | TCAAGAAAGCCATCCCACCC | TGGCAGATCCAGTAAAGCGG |
| SlFAD6 | CTCTTATCACCACCGCCCTC | GCTGCATTCACGATTGGCAT |
| SlFAD14 | CTTCCGTTTCTGTTTCCGCC | TAAGCGGTGCACTTACCCTC |
| SlFAD18 | CACCTGGTCTGTGGGAGTTC | CCAGCCCCAGAAGTAAACCA |
| SlFAD21 | CGAGGTTGAGGTGCTTGACT | TTCCAGGTTGAGTCGGATGC |
| SlFAD23 | CCGTTACTTTTGCATGGCCT | CCCAAGCTAGCCCTTTCATC |
表1 RT-qPCR引物
Table 1 Primers for RT-qPCR
| 基因名称Gene name | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| Actin | ATCTTGGCTTCCCTCAGCAC | GCATCTCTGGTCCAGTAGGAAAT |
| SlFAD1 | TCTACGGTGTGCCCCTTCTA | AGTGATGGGTGAGTGTGCTG |
| SlFAD4 | TCAAGAAAGCCATCCCACCC | TGGCAGATCCAGTAAAGCGG |
| SlFAD6 | CTCTTATCACCACCGCCCTC | GCTGCATTCACGATTGGCAT |
| SlFAD14 | CTTCCGTTTCTGTTTCCGCC | TAAGCGGTGCACTTACCCTC |
| SlFAD18 | CACCTGGTCTGTGGGAGTTC | CCAGCCCCAGAAGTAAACCA |
| SlFAD21 | CGAGGTTGAGGTGCTTGACT | TTCCAGGTTGAGTCGGATGC |
| SlFAD23 | CCGTTACTTTTGCATGGCCT | CCCAAGCTAGCCCTTTCATC |
| 保守基序名称 Conservative motif name | 具体序列分布情况 Specific sequence distribution |
|---|---|
| Motif 1 | WVTAHECGHHAFSDYQWJBDTVGFILHSSJLVPYFSWKYSHRRHHSNTGS |
| Motif 2 | NKVFHNITDTHVLHHLFSTIPHYHAVEATKAIKPLLGEYYQFDGTPIYKA |
| Motif 3 | EVKKHNKAKDCWJIISGKVYDVTKFLDDHPGGDEVLLSATG |
| Motif 4 | RVPSSKPPFTJSDIKKAIPPHCFQRSLVRSFSYVVRDJVLVFILYYIAAT |
| Motif 5 | CIYGVPLLIVNGFIVLITYLHHTHASLPHYDSSEWDWLRGA |
| Motif 6 | NVSGRPYDRFASHYBPYSPJYNDRERLQI |
| Motif 7 | KDATDDFEDVGHSSSAREMLDKYYIGEIDSSTIPTKRKYTP |
| Motif 8 | IWRDFKECIYVEKDEESQDKGVFWYKNK |
| Motif 9 | PYNYLAWPIYWIAQGCVFTGI |
| Motif 10 | QPKGNDWFEKQTAGTIDIACSPQMDWFFGGLQFQLEHHLFPRLPRCQLRK |
表2 保守基序motif序列分布情况
Table 2 Distribution of conservative motif sequence
| 保守基序名称 Conservative motif name | 具体序列分布情况 Specific sequence distribution |
|---|---|
| Motif 1 | WVTAHECGHHAFSDYQWJBDTVGFILHSSJLVPYFSWKYSHRRHHSNTGS |
| Motif 2 | NKVFHNITDTHVLHHLFSTIPHYHAVEATKAIKPLLGEYYQFDGTPIYKA |
| Motif 3 | EVKKHNKAKDCWJIISGKVYDVTKFLDDHPGGDEVLLSATG |
| Motif 4 | RVPSSKPPFTJSDIKKAIPPHCFQRSLVRSFSYVVRDJVLVFILYYIAAT |
| Motif 5 | CIYGVPLLIVNGFIVLITYLHHTHASLPHYDSSEWDWLRGA |
| Motif 6 | NVSGRPYDRFASHYBPYSPJYNDRERLQI |
| Motif 7 | KDATDDFEDVGHSSSAREMLDKYYIGEIDSSTIPTKRKYTP |
| Motif 8 | IWRDFKECIYVEKDEESQDKGVFWYKNK |
| Motif 9 | PYNYLAWPIYWIAQGCVFTGI |
| Motif 10 | QPKGNDWFEKQTAGTIDIACSPQMDWFFGGLQFQLEHHLFPRLPRCQLRK |
| 基因名称Gene name | 脱落酸响应Abscisic acid response | 厌氧诱导响应Anaerobic induction | 生长素响应Auxin response | 昼夜节律调控Circadian control | 防御和应激响应 Defense and stress response | 干旱诱导响应Drought- inducibility | 低温响应Low-temperature response | 赤霉素响应Gibberellin- response | 分生组织表达Meristem expression | 水杨酸响应Salicylic acid response | 茉莉酸响应MeJA- response |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SlFAD1 | 5 | 4 | 0 | 0 | 1 | 1 | 2 | 0 | 1 | 2 | 8 |
| SlFAD2 | 0 | 5 | 1 | 0 | 1 | 2 | 1 | 0 | 1 | 0 | 6 |
| SlFAD3 | 4 | 1 | 0 | 1 | 1 | 1 | 4 | 2 | 0 | 1 | 8 |
| SlFAD4 | 8 | 3 | 2 | 1 | 2 | 1 | 0 | 0 | 0 | 0 | 6 |
| SlFAD5 | 13 | 3 | 2 | 0 | 1 | 2 | 0 | 2 | 1 | 1 | 6 |
| SlFAD6 | 1 | 3 | 2 | 1 | 1 | 4 | 0 | 2 | 1 | 2 | 8 |
| SlFAD7 | 9 | 5 | 2 | 0 | 1 | 2 | 1 | 3 | 2 | 2 | 10 |
| SlFAD8 | 8 | 4 | 2 | 0 | 0 | 2 | 3 | 2 | 0 | 2 | 6 |
| SlFAD9 | 4 | 2 | 0 | 0 | 1 | 1 | 1 | 2 | 1 | 2 | 6 |
| SlFAD10 | 4 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 4 |
| SlFAD11 | 3 | 3 | 1 | 0 | 0 | 0 | 0 | 2 | 1 | 3 | 4 |
| SlFAD12 | 3 | 1 | 0 | 1 | 2 | 0 | 2 | 1 | 1 | 3 | 6 |
| SlFAD13 | 8 | 2 | 1 | 1 | 0 | 1 | 3 | 3 | 1 | 0 | 0 |
| SlFAD14 | 5 | 3 | 1 | 0 | 2 | 5 | 2 | 4 | 1 | 0 | 4 |
| SlFAD15 | 4 | 2 | 3 | 0 | 1 | 1 | 0 | 1 | 0 | 2 | 8 |
| SlFAD16 | 11 | 2 | 2 | 0 | 0 | 0 | 1 | 2 | 1 | 4 | 16 |
| SlFAD17 | 3 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 |
| SlFAD18 | 1 | 6 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 6 |
| SlFAD19 | 14 | 1 | 5 | 1 | 1 | 1 | 0 | 1 | 4 | 0 | 24 |
| SlFAD20 | 10 | 7 | 1 | 1 | 2 | 0 | 3 | 2 | 0 | 0 | 2 |
| SlFAD21 | 5 | 2 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 6 |
| SlFAD22 | 5 | 1 | 0 | 3 | 1 | 0 | 1 | 3 | 2 | 2 | 6 |
| SlFAD23 | 4 | 3 | 1 | 1 | 0 | 0 | 2 | 2 | 0 | 2 | 10 |
| SlFAD24 | 3 | 6 | 0 | 3 | 2 | 2 | 3 | 0 | 0 | 2 | 8 |
| SlFAD25 | 1 | 4 | 2 | 1 | 2 | 1 | 0 | 2 | 2 | 1 | 4 |
| SlFAD26 | 0 | 6 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 2 | 8 |
表3 番茄SlFAD基因家族顺式作用元件功能及数量分析
Table 3 Analysis of promoter cis-acting elements of the tomato SlFAD gene family
| 基因名称Gene name | 脱落酸响应Abscisic acid response | 厌氧诱导响应Anaerobic induction | 生长素响应Auxin response | 昼夜节律调控Circadian control | 防御和应激响应 Defense and stress response | 干旱诱导响应Drought- inducibility | 低温响应Low-temperature response | 赤霉素响应Gibberellin- response | 分生组织表达Meristem expression | 水杨酸响应Salicylic acid response | 茉莉酸响应MeJA- response |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SlFAD1 | 5 | 4 | 0 | 0 | 1 | 1 | 2 | 0 | 1 | 2 | 8 |
| SlFAD2 | 0 | 5 | 1 | 0 | 1 | 2 | 1 | 0 | 1 | 0 | 6 |
| SlFAD3 | 4 | 1 | 0 | 1 | 1 | 1 | 4 | 2 | 0 | 1 | 8 |
| SlFAD4 | 8 | 3 | 2 | 1 | 2 | 1 | 0 | 0 | 0 | 0 | 6 |
| SlFAD5 | 13 | 3 | 2 | 0 | 1 | 2 | 0 | 2 | 1 | 1 | 6 |
| SlFAD6 | 1 | 3 | 2 | 1 | 1 | 4 | 0 | 2 | 1 | 2 | 8 |
| SlFAD7 | 9 | 5 | 2 | 0 | 1 | 2 | 1 | 3 | 2 | 2 | 10 |
| SlFAD8 | 8 | 4 | 2 | 0 | 0 | 2 | 3 | 2 | 0 | 2 | 6 |
| SlFAD9 | 4 | 2 | 0 | 0 | 1 | 1 | 1 | 2 | 1 | 2 | 6 |
| SlFAD10 | 4 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 4 |
| SlFAD11 | 3 | 3 | 1 | 0 | 0 | 0 | 0 | 2 | 1 | 3 | 4 |
| SlFAD12 | 3 | 1 | 0 | 1 | 2 | 0 | 2 | 1 | 1 | 3 | 6 |
| SlFAD13 | 8 | 2 | 1 | 1 | 0 | 1 | 3 | 3 | 1 | 0 | 0 |
| SlFAD14 | 5 | 3 | 1 | 0 | 2 | 5 | 2 | 4 | 1 | 0 | 4 |
| SlFAD15 | 4 | 2 | 3 | 0 | 1 | 1 | 0 | 1 | 0 | 2 | 8 |
| SlFAD16 | 11 | 2 | 2 | 0 | 0 | 0 | 1 | 2 | 1 | 4 | 16 |
| SlFAD17 | 3 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 |
| SlFAD18 | 1 | 6 | 1 | 1 | 1 | 2 | 2 | 2 | 1 | 1 | 6 |
| SlFAD19 | 14 | 1 | 5 | 1 | 1 | 1 | 0 | 1 | 4 | 0 | 24 |
| SlFAD20 | 10 | 7 | 1 | 1 | 2 | 0 | 3 | 2 | 0 | 0 | 2 |
| SlFAD21 | 5 | 2 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 6 |
| SlFAD22 | 5 | 1 | 0 | 3 | 1 | 0 | 1 | 3 | 2 | 2 | 6 |
| SlFAD23 | 4 | 3 | 1 | 1 | 0 | 0 | 2 | 2 | 0 | 2 | 10 |
| SlFAD24 | 3 | 6 | 0 | 3 | 2 | 2 | 3 | 0 | 0 | 2 | 8 |
| SlFAD25 | 1 | 4 | 2 | 1 | 2 | 1 | 0 | 2 | 2 | 1 | 4 |
| SlFAD26 | 0 | 6 | 0 | 0 | 0 | 1 | 0 | 2 | 0 | 2 | 8 |
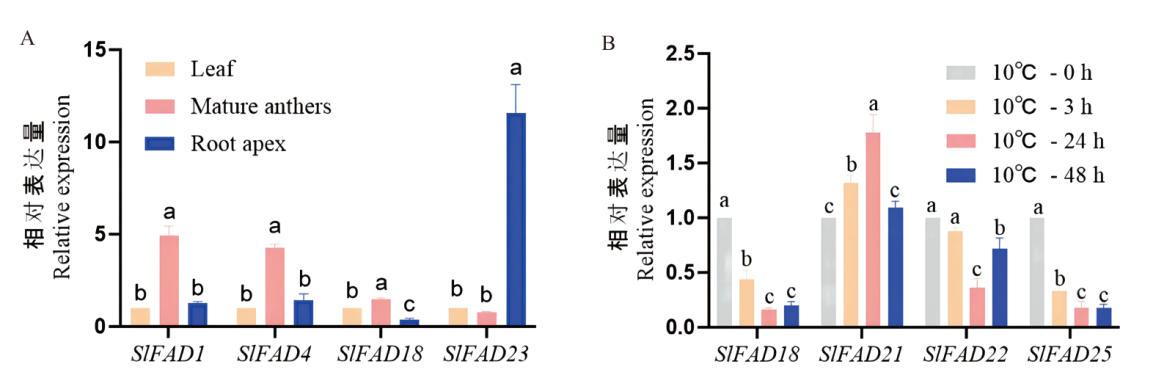
图6 不同组织及10℃下番茄SlFAD基因家族成员表达模式分析 不同字母表示处理间在0.05水平差异显著
Fig. 6 Analysis of expression patterns of tomato SlFAD gene family members in different tissues and at 10℃ Different letters indicate significant differences between treatments at the 0.05 level
| [1] |
Li NN, Xu CC, Li-Beisson Y, et al. Fatty acid and lipid transport in plant cells[J]. Trends Plant Sci, 2016, 21(2): 145-158.
doi: S1360-1385(15)00259-9 pmid: 26616197 |
| [2] | Cheng CZ, Liu F, Sun XL, et al. Genome-wide identification of FAD gene family and their contributions to the temperature stresses and mutualistic and parasitic fungi colonization responses in banana[J]. Int J Biol Macromol, 2022, 204: 661-676. |
| [3] | 李凤, 郑汪东, 唐文博. 谷子膜结合脂肪酸脱氢酶基因家族的鉴定及进化分析[J]. 山西农业科学, 2019, 47(7): 1121-1125. |
| Li F, Zheng WD, Tang WB. Identification and evolutionary analysis of membrane-bound fatty acid desaturases gene family in Setaria italica[J]. J Shanxi Agric Sci, 2019, 47(7): 1121-1125. | |
| [4] | 崔景焱. 亚麻芥中脂肪酸脱氢酶基因克隆分析与功能验证[D]. 长春: 吉林农业大学, 2019. |
| Cui JY. Cloning and functional characterization of fatty acid dehydrogenase gene in Camelina sativa[D]. Changchun: Jilin Agricultural University, 2019. | |
| [5] | 朱宗文, 张爱冬, 吴雪霞, 等. 生物信息学鉴定分析茄子脂肪酸去饱和酶(FAD)基因家族[J]. 分子植物育种, 2023, 21(8): 2453-2463. |
| Zhu ZW, Zhang AD, Wu XX, et al. Identification and bioinformatics analysis of fatty acid desaturase(FAD)gene family in eggplant(Solanum melongena L.)[J]. Mol Plant Breed, 2023, 21(8): 2453-2463. | |
| [6] |
焦苏淇, 周俊名, 尚雨晴, 等. 大豆脂肪酸脱氢酶GmFAD3C-1基因的克隆及功能分析[J]. 中国油料作物学报, 2022, 44(5): 1006-1017.
doi: 10.19802/j.issn.1007-9084.2021253 |
| Jiao SQ, Zhou JM, Shang YQ, et al. Cloning and genetic transformation of soybean fatty acid dehydrogenase GmFAD3C-1 gene[J]. Chin J Oil Crop Sci, 2022, 44(5): 1006-1017. | |
| [7] | 侯静静, 赵利, 王斌. 亚麻FAD基因家族的生物信息学鉴定分析[J]. 寒旱农业科学, 2023, 2(3): 246-253. |
| Hou JJ, Zhao L, Wang B. Identification and bioinformatics analysis of FAD gene family in Linum usitatissimum L[J]. J Cold Arid Agric Sci, 2023, 2(3): 246-253. | |
| [8] | 于海彦, 周志峰, 贾俊强, 等. 家蚕类ω3-脂肪酸脱氢酶基因的克隆、表达及功能研究[J]. 蚕业科学, 2017, 43(4): 577-586. |
| Yu HY, Zhou ZF, Jia JQ, et al. Cloning, expression and functional analysis of an ω3-fatty acid desaturase-like gene from Bombyx mori[J]. Sci Seric, 2017, 43(4): 577-586. | |
| [9] | 叶浩盈. 三角褐指藻脂肪酸去饱和酶基因家族分析及高产EPA藻株的选育[D]. 福州: 福建师范大学, 2019. |
| Ye HY. Analysis of fatty acid desaturase gene family of Phaeodactylum tricornutum and breeding of high-yield EPA algae strain[D]. Fuzhou: Fujian Normal University, 2019. | |
| [10] | 刘永红. 白沙蒿△12-脂肪酸脱氢酶基因克隆及其植物表达载体构建[D]. 兰州: 兰州大学, 2010. |
| Liu YH. Cloning and expression vector construction of △12-fatty acid desaturase(FAD2) gene from Artemisia sphaero-cephala[D]. Lanzhou: Lanzhou University, 2010. | |
| [11] |
Thiede MA, Ozols J, Strittmatter P. Construction and sequence of cDNA for rat liver stearyl coenzyme A desaturase[J]. J Biol Chem, 1986, 261(28): 13230-13235.
pmid: 2428815 |
| [12] | 缪秀梅. 白沙蒿FAD2基因家族克隆与功能研究及转基因苜蓿评价[D]. 兰州: 兰州大学, 2020. |
| Miao XM. Cloning and functional analysis of FAD2 gene family of Artemisia sphaerocephala and evaluation of transgenic alfalfa[D]. Lanzhou: Lanzhou University, 2020. | |
| [13] |
薛晓梦, 李建国, 白冬梅, 等. 花生FAD2基因家族表达分析及其对低温胁迫的响应[J]. 作物学报, 2019, 45(10): 1586-1594.
doi: 10.3724/SP.J.1006.2019.84177 |
| Xue XM, Li JG, Bai DM, et al. Expression profiles of FAD2 genes and their responses to cold stress in peanut[J]. Acta Agron Sin, 2019, 45(10): 1586-1594. | |
| [14] | Lou Y, Schwender J, Shanklin J. FAD2 and FAD3 desaturases form heterodimers that facilitate metabolic channeling in vivo[J]. J Biol Chem, 2014, 289(26): 17996-18007. |
| [15] | 李丹丹, 王庆, 胡博, 等. 低温对红花脂肪酸组成和脂肪酸脱氢酶基因表达的影响[J]. 分子植物育种, 2018, 16(16): 5223-5231. |
| Li DD, Wang Q, Hu B, et al. Effects of low temperature on fatty acid composition and gene expression of fatty acid desaturase in safflower[J]. Mol Plant Breed, 2018, 16(16): 5223-5231. | |
| [16] |
常培培, 张静, 杨建华, 等. 紫色番茄果实挥发性风味物质分析[J]. 食品科学, 2014, 35(14): 165-169.
doi: 10.7506/spkx1002-6630-201414032 |
| Chang PP, Zhang J, Yang JH, et al. Analysis of volatile compounds in purple tomato fruits[J]. Food Sci, 2014, 35(14): 165-169. | |
| [17] | 宋慧慧, 颜龙飞, 苟莎莎, 等. PuFAD8基因过表达载体的构建及其在番茄中的遗传转化[J]. 合肥工业大学学报: 自然科学版, 2023, 46(3): 392-396. |
| Song HH, Yan LF, Gou SS, et al. Construction of overexpression vector of PuFAD8 gene and its genetic transformation in tomato(Solanum lycopersicum)[J]. J Hefei Univ Technol Nat Sci, 2023, 46(3): 392-396. | |
| [18] | Wang JJ, Liu HR, Gao J, et al. Two ω-3 FADs are associated with peach fruit volatile formation[J]. International journal of molecular sciences, 2016, 17(4): 464. |
| [19] | 马银银, 李莉娜, 刘琦, 等. 苹果香气物质的组成、影响因素及调控策略[J]. 北方园艺, 2023(1): 119-127. |
| Ma YY, Li LN, Liu Q, et al. Composition, influencing factors and regulation strategies of aroma substances in apple[J]. North Hortic, 2023(1): 119-127. | |
| [20] | 赵训超, 魏玉磊, 丁冬, 等. 甜荞麦脂肪酸脱氢酶基因(FeFAD)家族的鉴定与分析[J]. 东北农业科学, 2021, 46(1): 36-41. |
| Zhao XC, Wei YL, Ding D, et al. Genome-wide identification and bioinformatics analysis of fatty acid desaturase gene(FeFAD)family in common buckwheat[J]. J Northeast Agric Sci, 2021, 46(1): 36-41. | |
| [21] | 向妮艳. 红花膜结合脂肪酸脱氢酶基因家族的鉴定及表达分析[D]. 武汉: 中南民族大学, 2020. |
| Xiang NY. Identification and expression analysis of membrane-bound fatty acid desaturases gene family in safflower[D]. Wuhan: South-central University for Nationalities, 2020. | |
| [22] | Barrero-Gil J, Huertas R, Rambla JL, et al. Tomato plants increase their tolerance to low temperature in a chilling acclimation process entailing comprehensive transcriptional and metabolic adjustments[J]. Plant Cell Environ, 2016, 39(10): 2303-2318. |
| [23] | Penin AA, Klepikova AV, Kasianov AS, et al. Comparative analysis of developmental transcriptome maps of Arabidopsis thaliana and Solanum lycopersicum[J]. Genes, 2019, 10(1): 50. |
| [24] | 包鹏. 续随子ω-3脂肪酸脱氢酶基因ElFAD3的克隆和功能研究[D]. 太谷: 山西农业大学, 2022. |
| Bao P. Cloning and functional analysis of ω-3 fatty acid dehydrogenase ElFAD3 gene in Euphorbia lathyris[D]. Taigu: Shanxi Agricultural University, 2022. | |
| [25] | 章妮, 王欢, 李绍峰, 等. 毛竹FAD基因家族鉴定与综合分析[J/OL]. 分子植物育种, 2023: 1-17. http://kns.cnki.net/kcms/detail/46.1068.S.20230309.1448.004.html. |
| Zhang N, Wang H, Li SF, et al. Identification and comprehensive analysis of FAD gene family of Bamboo[J/OL]. Molecular Plant Breeding, 2023:1-17. http://kns.cnki.net/kcms/detail/46.1068.S.20230309.1448.004.html. | |
| [26] | 冯吉妤. 陆地棉脂肪酸脱氢酶基因家族的全基因组鉴定和表达特征分析[D]. 杭州: 浙江大学, 2017. |
| Feng JY. Genome-wide identification of fatty acid desaturase gene family in Gossypium hirsutum L. and their expression patterns[D]. Hangzhou: Zhejiang University, 2017. | |
| [27] | Zhao XC, Wei JP, He L, et al. Identification of fatty acid desaturases in maize and their differential responses to low and high temperature[J]. Genes, 2019, 10(6): 445. |
| [28] | 阮建, 单雷, 李新国, 等. 花生FAD基因家族的全基因组鉴定与表达模式分析[J]. 山东农业科学, 2018, 50(6): 1-9. |
| Ruan J, Shan L, Li XG, et al. Genome-wide identification and expression pattern analysis of peanut FAD gene family[J]. Shandong Agric Sci, 2018, 50(6): 1-9. | |
| [29] | 陈义珍, 李浩, 傅明川, 等. 陆地棉膜结合脂肪酸去饱和酶基因家族的全基因组鉴定及表达分析[J]. 山东农业科学, 2023, 55(4): 11-23. |
| Chen YZ, Li H, Fu MC, et al. Genome-wide identification and expression analysis of membrane-bound fatty acid desaturase gene family in upland cotton(Gossypium hirsutum L.)[J]. Shandong Agric Sci, 2023, 55(4): 11-23. | |
| [30] | 金兄莲, 安进宝, 祁得胜, 等. 毛白杨FAD基因家族鉴定与表达模式分析[J/OL]. 分子植物育种 2022: 1-23. http://kns.cnki.net/kcms/detail/46.1068.S.20220825.1145.006.html. |
| Jin XL, An JB, Qi DS, et al. Identification and expression pattern analysis of FAD gene family in Populus tomentosa[J/OL]. Molecular Plant Breeding 2022: 1-23. http://kns.cnki.net/kcms/detail/46.1068.S.20220825.1145.006.html. |
| [1] | 臧文蕊, 马明, 砗根, 哈斯阿古拉. 甜瓜BZR转录因子家族基因的全基因组鉴定及表达模式分析[J]. 生物技术通报, 2024, 40(7): 163-171. |
| [2] | 李博静, 郑腊梅, 吴乌云, 高飞, 周宜君. 西蒙得木HSP20基因家族的进化、表达和功能分析[J]. 生物技术通报, 2024, 40(6): 190-202. |
| [3] | 王健, 杨莎, 孙庆文, 陈宏宇, 杨涛, 黄园. 金钗石斛bHLH转录因子家族全基因组鉴定及表达分析[J]. 生物技术通报, 2024, 40(6): 203-218. |
| [4] | 李梦然, 叶伟, 李赛妮, 张维阳, 李建军, 章卫民. Lithocarols类化合物生物合成基因litI的表达及其启动子功能分析[J]. 生物技术通报, 2024, 40(6): 310-318. |
| [5] | 王玉书, 赵琳琳, 赵爽, 胡琦, 白慧霞, 王欢, 曹业萍, 范震宇. 大白菜BrCYP83B1基因的克隆及表达分析[J]. 生物技术通报, 2024, 40(6): 152-160. |
| [6] | 胡永波, 雷雨田, 杨永森, 陈馨, 林黄昉, 林碧英, 刘爽, 毕格, 申宝营. 黄瓜和南瓜Bcl-2相关抗凋亡家族全基因组鉴定与表达模式分析[J]. 生物技术通报, 2024, 40(6): 219-237. |
| [7] | 常雪瑞, 王田田, 王静. 辣椒E2基因家族的鉴定及分析[J]. 生物技术通报, 2024, 40(6): 238-250. |
| [8] | 刘蓉, 田闵玉, 李光泽, 谭成方, 阮颖, 刘春林. 甘蓝型油菜REVEILLE家族鉴定及诱导表达分析[J]. 生物技术通报, 2024, 40(6): 161-171. |
| [9] | 郝思怡, 张君珂, 王斌, 曲朋燕, 李瑞得, 程春振. 香蕉ELF3的克隆与表达分析[J]. 生物技术通报, 2024, 40(5): 131-140. |
| [10] | 侯雅琼, 郎红珊, 闻蒙蒙, 谷易云, 朱润洁, 汤晓丽. 猕猴桃AcHSP20基因家族的鉴定及表达分析[J]. 生物技术通报, 2024, 40(5): 167-178. |
| [11] | 李嘉欣, 李鸿燕, 刘丽娥, 张恬, 周武. 沙棘NRAMP基因家族鉴定及铅胁迫下表达分析[J]. 生物技术通报, 2024, 40(5): 191-202. |
| [12] | 娄银, 高浩竣, 王茜, 牛景萍, 王敏, 杜维俊, 岳爱琴. 大豆GmHMGS基因的鉴定及表达模式分析[J]. 生物技术通报, 2024, 40(4): 110-121. |
| [13] | 陈春林, 李白雪, 李金玲, 杜清洁, 李猛, 肖怀娟. 甜瓜CmEPF基因家族的鉴定及表达分析[J]. 生物技术通报, 2024, 40(4): 130-138. |
| [14] | 肖雅茹, 贾婷婷, 罗丹, 武喆, 李丽霞. 黄瓜CsERF025L转录因子的克隆及表达分析[J]. 生物技术通报, 2024, 40(4): 159-166. |
| [15] | 钟匀, 林春, 刘正杰, 董陈文华, 毛自朝, 李兴玉. 芦笋皂苷合成相关糖基转移酶基因克隆及原核表达分析[J]. 生物技术通报, 2024, 40(4): 255-263. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||