








生物技术通报 ›› 2024, Vol. 40 ›› Issue (9): 123-130.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0511
• 薯类作物生物技术专题(专题主编:徐建飞,尚轶) • 上一篇 下一篇
夏士轩1,2( ), 耿泽栋1, 祝光涛3, 张春芝2, 李大伟2(
), 耿泽栋1, 祝光涛3, 张春芝2, 李大伟2( )
)
收稿日期:2024-05-29
出版日期:2024-09-26
发布日期:2024-10-12
通讯作者:
李大伟,男,博士,助理研究员,研究方向:马铃薯分子育种;E-mail: lidawei@caas.cn作者简介:夏士轩,男,硕士,研究方向:马铃薯育性分子机制;E-mail: xiaxuan172@163.com
基金资助:
XIA Shi-xuan1,2( ), GENG Ze-dong1, ZHU Guang-tao3, ZHANG Chun-zhi2, LI Da-wei2(
), GENG Ze-dong1, ZHU Guang-tao3, ZHANG Chun-zhi2, LI Da-wei2( )
)
Received:2024-05-29
Published:2024-09-26
Online:2024-10-12
摘要:
【目的】传统马铃薯花粉活力检测方法依靠肉眼计数,存在效率低、准确性差等问题。本研究基于PaddlePaddle深度学习框架,通过比较不同模型,提出一种快速检测花粉活力的方法。【方法】首先收集花粉,使用2,3,5-氯化三苯基四氮唑(2,3,5-triphenyltetrazolium chloride,TTC)染色,通过显微镜拍照获取图像;利用Photoshop(PS)进行数据标注,分别标注有活力花粉和所有花粉,并将标签图转为单通道图像;选用SegFormer、U-Net和DeepLabV3模型进行训练,分割有活力花粉和所有花粉;最后使用Python OpenCV程序计数,计算花粉活力。【结果】与其他模型相比,SegFormer在两类数据集中的各项评估指标均为最优。相比于人工识别,OpenCV程序可以实现快速、批量计数,且误差小。【结论】通过图像处理技术可以快速、准确检测马铃薯花粉活力,进一步利用该方法快速鉴定了200份F2株系的花粉活力,为马铃薯花粉活力表型采集奠定了基础。
夏士轩, 耿泽栋, 祝光涛, 张春芝, 李大伟. 基于深度学习的马铃薯花粉活力快速检测[J]. 生物技术通报, 2024, 40(9): 123-130.
XIA Shi-xuan, GENG Ze-dong, ZHU Guang-tao, ZHANG Chun-zhi, LI Da-wei. Quick Detection of Potato Pollen Viability Based on Deep Learning[J]. Biotechnology Bulletin, 2024, 40(9): 123-130.
| 数据集 Dataset | 模型 Model | 交并比 IoU | 精确率 Precision | 召回率 Recall | F1分数 F1 score |
|---|---|---|---|---|---|
| 有活力花粉 | SegFormer | 35.25 | 85.95 | 38.26 | 51.11 |
| U-Net | 34.34 | 78.62 | 38.28 | 49.92 | |
| DeepLabV3 | 31.34 | 78.19 | 35.04 | 47.35 | |
| 所有花粉 | SegFormer | 75.65 | 82.67 | 88.64 | 84.78 |
| U-Net | 58.50 | 74.56 | 72.58 | 73.08 | |
| DeepLabV3 | 59.08 | 78.26 | 69.97 | 73.70 |
表1 不同模型对比实验的结果
Table 1 Comparative results of different algorithms
| 数据集 Dataset | 模型 Model | 交并比 IoU | 精确率 Precision | 召回率 Recall | F1分数 F1 score |
|---|---|---|---|---|---|
| 有活力花粉 | SegFormer | 35.25 | 85.95 | 38.26 | 51.11 |
| U-Net | 34.34 | 78.62 | 38.28 | 49.92 | |
| DeepLabV3 | 31.34 | 78.19 | 35.04 | 47.35 | |
| 所有花粉 | SegFormer | 75.65 | 82.67 | 88.64 | 84.78 |
| U-Net | 58.50 | 74.56 | 72.58 | 73.08 | |
| DeepLabV3 | 59.08 | 78.26 | 69.97 | 73.70 |

图2 不同尺度图片的分割效果 原图为花粉TTC染色图像;伪彩色图为模型输出的花粉分割图像;掩膜图是为方便观察分割效果,将伪彩色图与原图掩膜后的结果;200 μm(左图);400 μm(右图)
Fig. 2 Segmentation results of different-scale pictures The original image is the pollen stained by TTC; the pseudo-color image is the pollen segmentation image output by the model; the mask image is the result of masking the pseudo-color image and the original image for the convenience of observing the segmentation effect; 200 μm(left); 400 μm(right)
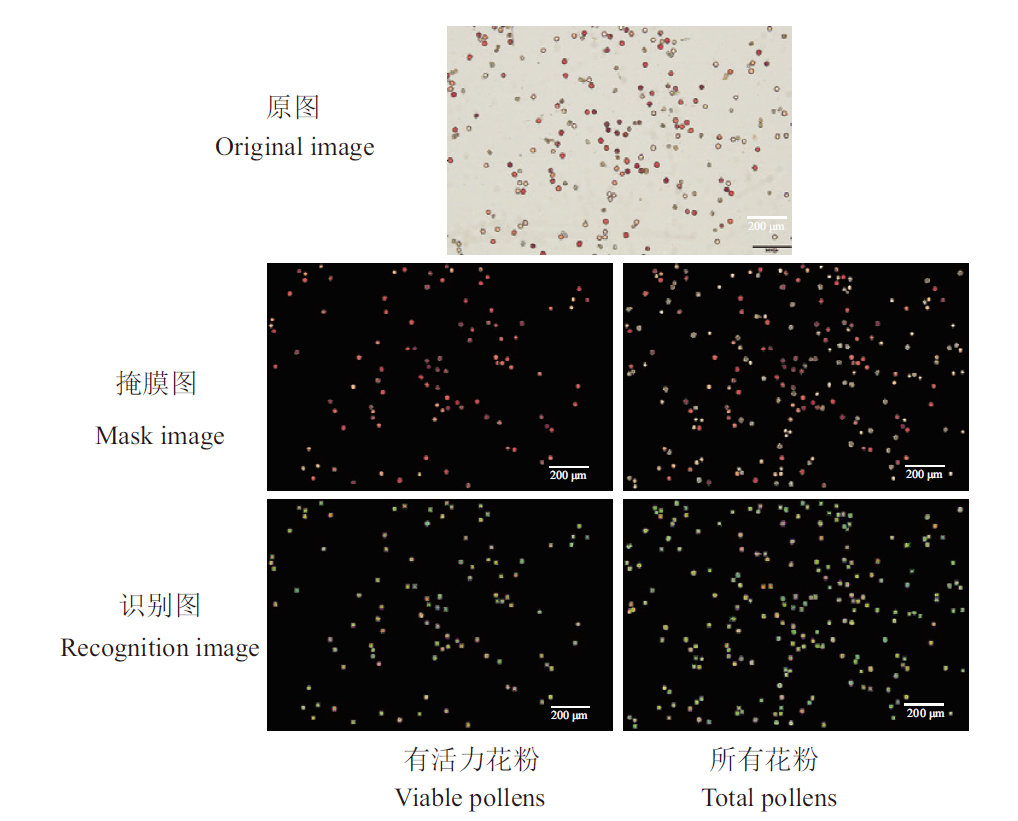
图3 OpenCV程序识别效果 原图为花粉TTC染色图像;掩膜图为模型输出的伪彩色图与原图掩膜的结果;识别图为OpenCV程序对识别到的花粉进行标记后输出的图像;比例尺200 μm
Fig. 3 Recognition effect by OpenCV program The original image is the pollen stained by TTC; the mask image is the result of masking the pseudo-color image output by the model with the original image; the recognition image is the output by the OpenCV program after labeling the recognized pollens; bar = 200 μm
| 编号 No. | 有活力花粉数The number of viable pollens | 总花粉数The number of total pollens | 花粉活力Pollen viability | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 程序计算值Calculated value by program | 人工计算值Manually calculated value | 误差 Error | 程序计算值Calculated value by program | 人工计算值Manually calculated value | 误差 Error | 程序计算值Calculated value by program/% | 人工计算值Manually calculated value/% | 误差 Error/% | ||||
| 2×-1 | 569 | 535 | -34 | 1 074 | 1 021 | -53 | 53 | 52 | -1 | |||
| 2×-2 | 241 | 240 | -1 | 442 | 457 | 15 | 55 | 53 | -2 | |||
| 2×-3 | 297 | 296 | -1 | 390 | 403 | 13 | 76 | 73 | -3 | |||
| 2×-4 | 447 | 445 | -2 | 528 | 553 | 25 | 84 | 80 | -4 | |||
| 2×-5 | 355 | 357 | 2 | 483 | 511 | 28 | 73 | 70 | -3 | |||
| 2×-6 | 158 | 154 | -4 | 315 | 303 | -12 | 50 | 51 | 1 | |||
| 2×-7 | 99 | 103 | 4 | 138 | 153 | 15 | 72 | 68 | -4 | |||
| 2×-8 | 167 | 161 | -6 | 235 | 238 | 3 | 71 | 68 | -3 | |||
| 2×-9 | 142 | 139 | -3 | 177 | 183 | 6 | 80 | 76 | -4 | |||
| 2×-10 | 314 | 314 | 0 | 553 | 537 | -16 | 57 | 58 | 1 | |||
| 4×-1 | 117 | 125 | 8 | 249 | 270 | 21 | 47 | 46 | -1 | |||
| 4×-2 | 118 | 124 | 6 | 244 | 263 | 19 | 48 | 47 | -1 | |||
| 4×-3 | 80 | 82 | 2 | 174 | 180 | 6 | 46 | 46 | 0 | |||
| 4×-4 | 71 | 70 | -1 | 187 | 199 | 12 | 38 | 35 | -3 | |||
| 4×-5 | 72 | 79 | 7 | 240 | 246 | 6 | 30 | 32 | 2 | |||
| 4×-6 | 73 | 81 | 8 | 245 | 257 | 12 | 30 | 32 | 2 | |||
| 4×-7 | 70 | 74 | 4 | 204 | 218 | 14 | 34 | 34 | 0 | |||
| 4×-8 | 75 | 73 | -2 | 188 | 206 | 18 | 40 | 35 | -4 | |||
| 4×-9 | 69 | 75 | 6 | 232 | 251 | 19 | 30 | 30 | 0 | |||
| 4×-10 | 56 | 60 | 4 | 186 | 198 | 12 | 30 | 30 | 0 | |||
表2 不同识别方法的结果对比
Table 2 Comparison of different identification methods
| 编号 No. | 有活力花粉数The number of viable pollens | 总花粉数The number of total pollens | 花粉活力Pollen viability | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 程序计算值Calculated value by program | 人工计算值Manually calculated value | 误差 Error | 程序计算值Calculated value by program | 人工计算值Manually calculated value | 误差 Error | 程序计算值Calculated value by program/% | 人工计算值Manually calculated value/% | 误差 Error/% | ||||
| 2×-1 | 569 | 535 | -34 | 1 074 | 1 021 | -53 | 53 | 52 | -1 | |||
| 2×-2 | 241 | 240 | -1 | 442 | 457 | 15 | 55 | 53 | -2 | |||
| 2×-3 | 297 | 296 | -1 | 390 | 403 | 13 | 76 | 73 | -3 | |||
| 2×-4 | 447 | 445 | -2 | 528 | 553 | 25 | 84 | 80 | -4 | |||
| 2×-5 | 355 | 357 | 2 | 483 | 511 | 28 | 73 | 70 | -3 | |||
| 2×-6 | 158 | 154 | -4 | 315 | 303 | -12 | 50 | 51 | 1 | |||
| 2×-7 | 99 | 103 | 4 | 138 | 153 | 15 | 72 | 68 | -4 | |||
| 2×-8 | 167 | 161 | -6 | 235 | 238 | 3 | 71 | 68 | -3 | |||
| 2×-9 | 142 | 139 | -3 | 177 | 183 | 6 | 80 | 76 | -4 | |||
| 2×-10 | 314 | 314 | 0 | 553 | 537 | -16 | 57 | 58 | 1 | |||
| 4×-1 | 117 | 125 | 8 | 249 | 270 | 21 | 47 | 46 | -1 | |||
| 4×-2 | 118 | 124 | 6 | 244 | 263 | 19 | 48 | 47 | -1 | |||
| 4×-3 | 80 | 82 | 2 | 174 | 180 | 6 | 46 | 46 | 0 | |||
| 4×-4 | 71 | 70 | -1 | 187 | 199 | 12 | 38 | 35 | -3 | |||
| 4×-5 | 72 | 79 | 7 | 240 | 246 | 6 | 30 | 32 | 2 | |||
| 4×-6 | 73 | 81 | 8 | 245 | 257 | 12 | 30 | 32 | 2 | |||
| 4×-7 | 70 | 74 | 4 | 204 | 218 | 14 | 34 | 34 | 0 | |||
| 4×-8 | 75 | 73 | -2 | 188 | 206 | 18 | 40 | 35 | -4 | |||
| 4×-9 | 69 | 75 | 6 | 232 | 251 | 19 | 30 | 30 | 0 | |||
| 4×-10 | 56 | 60 | 4 | 186 | 198 | 12 | 30 | 30 | 0 | |||
| [1] | Hardigan MA, Laimbeer FPE, Newton L, et al. Genome diversity of tuber-bearing Solanum uncovers complex evolutionary history and targets of domestication in the cultivated potato[J]. Proc Natl Acad Sci USA, 2017, 114(46): E9999-E10008. |
| [2] |
Camire ME, Kubow S, Donnelly DJ. Potatoes and human health[J]. Crit Rev Food Sci Nutr, 2009, 49(10): 823-840.
doi: 10.1080/10408390903041996 pmid: 19960391 |
| [3] | Monro J, Mishra S, Blandford E, et al. Potato genotype differences in nutritionally distinct starch fractions after cooking, and cooking plus storing cool[J]. J Food Compos Anal, 2009, 22(6): 539-545. |
| [4] | Aliche EB, Theeuwen TPJM, Oortwijn M, et al. Carbon partitioning mechanisms in potato under drought stress[J]. Plant Physiol Biochem, 2020, 146: 211-219. |
| [5] | Simakov EA, Anisimov BV, Yashina IM, et al. Potato breeding and seed production system development in Russia[J]. Potato Res, 2008, 51(3): 313-326. |
| [6] | Celis C, Scurrah M, Cowgill S, et al. Environmental biosafety and transgenic potato in a centre of diversity for this crop[J]. Nature, 2004, 432(7014): 222-225. |
| [7] |
Gebhardt C, Valkonen JP. Organization of genes controlling disease resistance in the potato genome[J]. Annu Rev Phytopathol, 2001, 39: 79-102.
pmid: 11701860 |
| [8] | Lindhout P, Meijer D, Schotte T, et al. Towards F1 hybrid seed potato breeding[J]. Potato Res, 2011, 54(4): 301-312. |
| [9] | Boavida LC, Vieira AM, Becker JD, et al. Gametophyte interaction and sexual reproduction: how plants make a zygote[J]. Int J Dev Biol, 2005, 49(5/6): 615-632. |
| [10] | Dafni A, Firmage D. Pollen viability and longevity: practical, ecological and evolutionary implications[J]. Plant Syst Evol, 2000, 222(1): 113-132. |
| [11] | Chen DJ, Neumann K, Friedel S, et al. Dissecting the phenotypic components of crop plant growth and drought responses based on high-throughput image analysis[J]. Plant Cell, 2014, 26(12): 4636-4655. |
| [12] |
Walter A, Liebisch F, Hund A. Plant phenotyping: from bean weighing to image analysis[J]. Plant Methods, 2015, 11: 14.
doi: 10.1186/s13007-015-0056-8 pmid: 25767559 |
| [13] |
Song P, Wang JL, Guo XY, et al. High-throughput phenotyping: breaking through the bottleneck in future crop breeding[J]. Crop J, 2021, 9(3): 633-645.
doi: 10.1016/j.cj.2021.03.015 |
| [14] | García-Fortea E, García-Pérez A, Gimeno-Páez E, et al. A deep learning-based system(microscan)for the identification of pollen development stages and its application to obtaining doubled haploid lines in eggplant[J]. Biology, 2020, 9(9): 272. |
| [15] | 周成全, 叶宏宝, 俞国红, 等. 基于机器视觉与深度学习的西兰花表型快速提取方法研究[J]. 智慧农业, 2020, 2(1): 121-132. |
|
Zhou CQ, Ye HB, Yu GH, et al. A fast extraction method of broccoli phenotype based on machine vision and deep learning[J]. Smart Agric, 2020, 2(1): 121-132.
doi: 10.12133/j.smartag.2020.2.1.201912-SA003 |
|
| [16] | 赵越, 赵辉, 姜永成, 等. 基于深度学习的马铃薯叶片病害检测方法[J]. 中国农机化学报, 2022, 43(10): 183-189. |
| Zhao Y, Zhao H, Jiang YC, et al. Detection method of potato leaf diseases based on deep learning[J]. J Chin Agric Mech, 2022, 43(10): 183-189. | |
| [17] | 胡松涛, 翟瑞芳, 王应华, 等. 基于多源数据的马铃薯植株表型参数提取[J]. 智慧农业, 2023, 5(1):132-145. |
|
Hu ST, Zhai RF, Wang YH, et al. Extraction of potato plant phenotypic parameters based on multi-source data[J]. Smart Agric, 2023, 5(1): 132-145.
doi: 10.12133/j.smartag.SA202302009 |
|
| [18] | Gao Y, Li YL, Jiang RB, et al. Enhancing green fraction estimation in rice and wheat crops: a self-supervised deep learning semantic segmentation approach[J]. Plant Phenomics, 2023, 5: 0064. |
| [19] |
Tello J, Montemayor MI, Forneck A, et al. A new image-based tool for the high throughput phenotyping of pollen viability: evaluation of inter- and intra-cultivar diversity in grapevine[J]. Plant Methods, 2018, 14: 3.
doi: 10.1186/s13007-017-0267-2 pmid: 29339970 |
| [20] | Tan ZH, Yang J, Li QY, et al. PollenDetect: an open-source pollen viability status recognition system based on deep learning neural networks[J]. Int J Mol Sci, 2022, 23(21): 13469. |
| [21] | 马艳军, 于佃海, 吴甜, 等. 飞桨:源于产业实践的开源深度学习平台[J]. 数据与计算发展前沿, 2019, 1(5):105-115. |
| Ma YJ, Yu DH, Wu T, et al. PaddlePaddle: an open-source deep learning platform from industrial practice[J]. Frontiers of Data and Computing, 2019, 1(5): 105-115. | |
| [22] | Hao SJ, Zhou Y, Guo YR. A brief survey on semantic segmentation with deep learning[J]. Neurocomputing, 2020, 406: 302-321. |
| [23] |
Zhang CZ, Yang ZM, Tang D, et al. Genome design of hybrid potato[J]. Cell, 2021, 184(15): 3873-3883.e12.
doi: 10.1016/j.cell.2021.06.006 pmid: 34171306 |
| [24] | Sun C, Shrivastava A, Singh S, et al. Revisiting unreasonable effectiveness of data in deep learning era[C]// 2017 IEEE International Conference on Computer Vision(ICCV). Venice, Italy. Piscataway, NJ: IEEE, 2017: 843-852. |
| [25] | Shorten C, Khoshgoftaar TM. A survey on image data augmentation for deep learning[J]. J Big Data, 2019, 6(1): 60. |
| [26] | Su D, Kong H, Qiao YL, et al. Data augmentation for deep learning based semantic segmentation and crop-weed classification in agricultural robotics[J]. Comput Electron Agric, 2021, 190: 106418. |
| [27] | Dosovitskiy A, Beyer L, Kolesnikov A, et al. An image is worth 16x16 words: transformers for image recognition at scale[J]. arXiv E Prints, 2020: arXiv: 2010.11929. |
| [28] | Xie EZ, Wang WH, Yu ZD, et al. SegFormer: simple and efficient design for semantic segmentation with transformers[EB/OL]. 2021: arXiv: 2105. 15203. http://arxiv.org/abs/2105.15203. |
| [29] | Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation[C]// International Conference on Medical Image Computing and Computer-Assisted Intervention. Cham: Springer, 2015: 234-241. |
| [30] | Chen LC, Papandreou G, Schroff F, et al. Rethinking atrous convolution for semantic image segmentation[EB/OL]. 2017: arXiv: 1706.05587. http://arxiv.org/abs/1706.05587. |
| [31] |
Yang WN, Feng H, Zhang XH, et al. Crop phenomics and high-throughput phenotyping: past decades, current challenges, and future perspectives[J]. Mol Plant, 2020, 13(2): 187-214.
doi: S1674-2052(20)30008-3 pmid: 31981735 |
| [32] | 仇瑞承, 魏爽, 张漫, 等. 作物表型组学测量方法综述[J]. 中国农业文摘-农业工程, 2019, 31(1): 23-36, 55. |
| Qiu RC, Wei S, Zhang M, et al. Summary of crop phenotypic omics measurement methods[J]. Agric Sci Eng China, 2019, 31(1): 23-36, 55. |
| [1] | 申鹏, 高雅彬, 丁红. 马铃薯SAT基因家族的鉴定和表达分析[J]. 生物技术通报, 2024, 40(9): 64-73. |
| [2] | 宋兵芳, 柳宁, 程新艳, 徐晓斌, 田文茂, 高悦, 毕阳, 王毅. 马铃薯G6PDH基因家族鉴定及其在损伤块茎的表达分析[J]. 生物技术通报, 2024, 40(9): 104-112. |
| [3] | 王超, 白如仟, 管俊梅, 罗稷林, 何雪姣, 迟绍轶, 马玲. 马铃薯块茎变绿中StHY5对龙葵素合成的促进作用[J]. 生物技术通报, 2024, 40(9): 113-122. |
| [4] | 毛向红, 卢瑶, 范向斌, 杜培兵, 白小东. 基于SSR荧光标记毛细管电泳的马铃薯品种遗传多样性分析及分子身份证构建[J]. 生物技术通报, 2024, 40(9): 131-140. |
| [5] | 袁兰, 黄娅楠, 张贝妮, 熊雨萌, 王洪洋. 基于流式细胞仪鉴定马铃薯倍性的高通量样品制备方法[J]. 生物技术通报, 2024, 40(9): 141-147. |
| [6] | 宋倩娜, 段永红, 冯瑞云. CRISPR/Cas9介导的高效四倍体马铃薯试管薯基因编辑体系的建立[J]. 生物技术通报, 2024, 40(9): 33-41. |
| [7] | 王柯然, 闫俊杰, 刘建凤, 高玉林. RNAi技术在马铃薯害虫防控中的应用和风险[J]. 生物技术通报, 2024, 40(9): 4-10. |
| [8] | 张小妹, 周南伶, 张赛行, 王超, 沈玉龙, 管俊梅, 马玲. 马铃薯StDREBs基因的克隆及其表达分析[J]. 生物技术通报, 2024, 40(9): 42-50. |
| [9] | 满全财, 孟姿诺, 李伟, 蔡心汝, 苏润东, 付长青, 高顺娟, 崔江慧. 马铃薯AQP基因家族鉴定及表达分析[J]. 生物技术通报, 2024, 40(9): 51-63. |
| [10] | 吴娟, 武小娟, 王沛捷, 谢锐, 聂虎帅, 李楠, 马艳红. 彩色马铃薯花青素合成相关ERF基因筛选及表达分析[J]. 生物技术通报, 2024, 40(9): 82-91. |
| [11] | 乔岩, 杨芳, 任盼荣, 祁伟亮, 安沛沛, 李茜, 李丹, 肖俊飞. 马铃薯野生种烯酰水合酶超家族基因ScDHNS的克隆与功能分析[J]. 生物技术通报, 2024, 40(9): 92-103. |
| [12] | 张震, 李清, 徐菁, 陈凯园, 张春芝, 祝光涛. 马铃薯线粒体靶向表达载体的构建与应用[J]. 生物技术通报, 2024, 40(5): 66-73. |
| [13] | 张玉, 石磊, 巩檑, 聂峰杰, 杨江伟, 刘璇, 杨文静, 张国辉, 颉瑞霞, 张丽. 马铃薯WOX基因家族的鉴定及在离体再生和非生物胁迫中的表达分析[J]. 生物技术通报, 2024, 40(3): 170-180. |
| [14] | 梅显军, 宋慧洋, 李京昊, 梅超, 宋倩娜, 冯瑞云, 陈喜明. 马铃薯StDof5的克隆及表达分析[J]. 生物技术通报, 2024, 40(3): 181-192. |
| [15] | 张春芝, 周倩, 吴瑶瑶, 尚轶, 黄三文. 基因组学研究助力马铃薯育种方式的变革[J]. 生物技术通报, 2024, 40(10): 11-18. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||