








生物技术通报 ›› 2024, Vol. 40 ›› Issue (9): 190-197.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0321
崔海洋( ), 谭淼, 全壮, 陈红利, 董艳敏, 唐立春(
), 谭淼, 全壮, 陈红利, 董艳敏, 唐立春( )
)
收稿日期:2024-03-29
出版日期:2024-09-26
发布日期:2024-10-12
通讯作者:
唐立春,男,博士,副研究员,研究方向:生物化学与分子生物学;E-mail: tlc@taiyuanshengwu.com作者简介:崔海洋,男,硕士,研发助理,研究方向:生物化学与分子生物学;E-mail: chy@taiyuanshengwu.com
CUI Hai-yang( ), TAN Miao, QUAN Zhuang, CHEN Hong-li, DONG Yan-min, TANG Li-chun(
), TAN Miao, QUAN Zhuang, CHEN Hong-li, DONG Yan-min, TANG Li-chun( )
)
Received:2024-03-29
Published:2024-09-26
Online:2024-10-12
摘要:
【目的】Cas9TX是Cas9的变体,能够显著降低基因编辑过程中染色体易位,大幅提升基因编辑的安全性。研究Cas9TX替换Cas9实现基因定点敲入的可行性。【方法】首先,利用His标签蛋白纯化技术制备Cas9TX蛋白,并在细胞水平上通过绘制EC50和核酸酶裂解动力学曲线等进行功能验证。其次,以RNP和dsDNA混合物电转的方式编辑体外激活的T细胞,流式检测定点敲入的效率。最后,探讨了供体模板进行稳定性修饰提升定点敲入效率的可行性。【结果】制备的Cas9TX在T细胞TRAC基因的A、R和S靶点的敲除效率分别为71.8%、81.0%和79.9%,具有与Cas9相当的基因敲除效率。在TRAC 1号外显子分别设计的dsDNA供体模板(2A-GFP编码序列)和ssDNA供体模板(+ GTC bp),发现Cas9TX RNP定点敲入两种模板的效率均显著低于Cas9 RNP。通过DNA修饰制备防TREX2核酸外切酶降解的供体模板,不能提升Cas9TX RNP 定点敲入的效率。【结论】Cas9TX RNP在TRAC三个靶点的基因敲除方面可以替代Cas9 RNP使用,但定点整合外源基因的效率约为Cas9 RNP的一半。为Cas9TX应用于基因定点敲入提供了重要参考。
崔海洋, 谭淼, 全壮, 陈红利, 董艳敏, 唐立春. 利用Cas9TX实现非病毒TRAC定点整合制备T细胞[J]. 生物技术通报, 2024, 40(9): 190-197.
CUI Hai-yang, TAN Miao, QUAN Zhuang, CHEN Hong-li, DONG Yan-min, TANG Li-chun. Generation of Virus-free TRAC-knocked-in T Cells Using Cas9TX[J]. Biotechnology Bulletin, 2024, 40(9): 190-197.
| 名称Name | 序列Sequence(5'-3') | |
|---|---|---|
| PCR Primers[ | TRAC-F4 | ATCACGAGCAGCTGGTTTCT |
| TRAC-R3 | GCCACCTTCTCTTCATCTGC | |
| qPCR Primers[ | TRAC-Up-F | GCATTTCAGGTTTCCTTGAGTGGCAG |
| TRAC-Up-R | TGGCAAGTCACGGTCTCATGCTTTAT | |
| TRAC-Cross-F | CTTGTCCATCACTGGCATCTGGACTC | |
| TRAC-Cross-R | ATCGGTGTGAATAGGCAGACAGACTTGT | |
| gRNAs[ | TRAC-A | ACAGATATCCAGAACCCTG |
| TRAC-R | TGTACCAGCTGAGAGACTCT | |
| TRAC-S | ACAAAACTGTGCTAGACATG | |
| ssDNA Templates[ | ssODN-F/ssODN-3'P | AGTAAGGATTCTGATGTGTATATCACAGACAAAACTGTGCTAGTCGACATGAGGTCTATGGACTTCAAGAGCAACAGTGCTGTGGCCTGGAGC |
| ssODN-R/ssODN-R-3'P | GCTCCAGGCCACAGCACTGTTGCTCTTGAAGTCCATAGACCTCATGTCGACTAGCACAGTTTTGTCTGTGATATACACATCAGAATCCTTACT | |
表1 序列信息
Table 1 Sequences information
| 名称Name | 序列Sequence(5'-3') | |
|---|---|---|
| PCR Primers[ | TRAC-F4 | ATCACGAGCAGCTGGTTTCT |
| TRAC-R3 | GCCACCTTCTCTTCATCTGC | |
| qPCR Primers[ | TRAC-Up-F | GCATTTCAGGTTTCCTTGAGTGGCAG |
| TRAC-Up-R | TGGCAAGTCACGGTCTCATGCTTTAT | |
| TRAC-Cross-F | CTTGTCCATCACTGGCATCTGGACTC | |
| TRAC-Cross-R | ATCGGTGTGAATAGGCAGACAGACTTGT | |
| gRNAs[ | TRAC-A | ACAGATATCCAGAACCCTG |
| TRAC-R | TGTACCAGCTGAGAGACTCT | |
| TRAC-S | ACAAAACTGTGCTAGACATG | |
| ssDNA Templates[ | ssODN-F/ssODN-3'P | AGTAAGGATTCTGATGTGTATATCACAGACAAAACTGTGCTAGTCGACATGAGGTCTATGGACTTCAAGAGCAACAGTGCTGTGGCCTGGAGC |
| ssODN-R/ssODN-R-3'P | GCTCCAGGCCACAGCACTGTTGCTCTTGAAGTCCATAGACCTCATGTCGACTAGCACAGTTTTGTCTGTGATATACACATCAGAATCCTTACT | |
| 组分Components | 用量Volume/μL |
|---|---|
| Nuclease Free ddH2O | 19.8 |
| NEB Buffer 2.1 | 2.5 |
| TRAC DNA Template | 2 |
| RNP | 0.7 |
| Total | 25 |
表2 RNP体外酶切反应体系
Table 2 Enzymatic digestion reaction system of RNP in vitro
| 组分Components | 用量Volume/μL |
|---|---|
| Nuclease Free ddH2O | 19.8 |
| NEB Buffer 2.1 | 2.5 |
| TRAC DNA Template | 2 |
| RNP | 0.7 |
| Total | 25 |
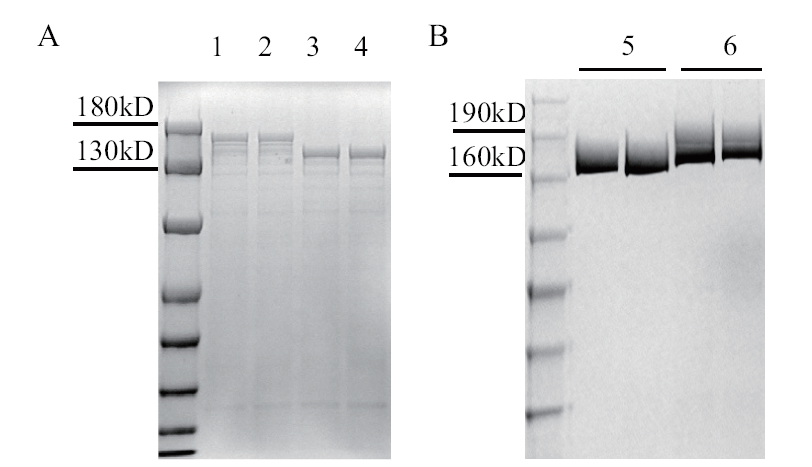
图1 SDS-PAGE检测Cas9和Cas9TX蛋白 1-2: Cas9TX 裂解液上清;3-4: Cas9 裂解液上清;5: 纯化的 Cas9 蛋白;6: 纯化的Cas9TX 蛋白
Fig. 1 Detection of Cas9 and Cas9TX protein by SDS-PAGE 1-2: Supernatant of Cas9TX bacterial lysate;3-4: supernatant of Cas9 bacterial lysate; 5: purified Cas9 protein; 6: purified Cas9TX protein
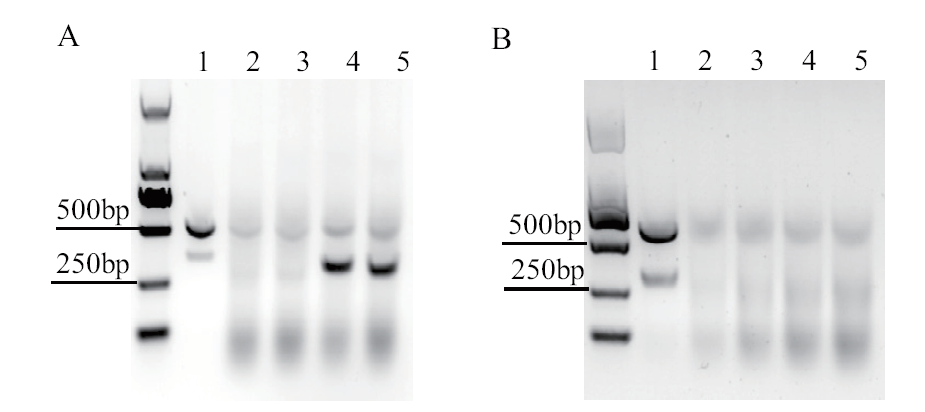
图2 Cas9和Cas9TX 体外酶切活性鉴定 A: RNP体外酶切目的片段. 1: 目的片段; 2-3: Cas9TX RNP+目的片段; 4-5: Cas9 RNP +目的片段; B: 不同浓度Cas9TX RNP切割目的片段. 1: 0.1 μmol/L; 2: 0.3 μmol/L; 3: 0.5 μmol/L; 4: 0.7 μmol/L; 5: 0.9 μmol/L
Fig. 2 Characterization of nuclease activity of Cas9 and Cas9TX in vitro A: RNP digestion of the TRAC DNA fragment. 1: target fragment; 2-3: target fragment with Cas9TX RNP; 4-5: target fragment with Cas9 RNP. B: Cut fragment with different concentrations of Cas9TX RNP. 1: 0.1 μmol/L; 2: 0.3 μmol/L; 3: 0.5 μmol/L; 4: 0.7 μmol/L; 5: 0.9 μmol/L
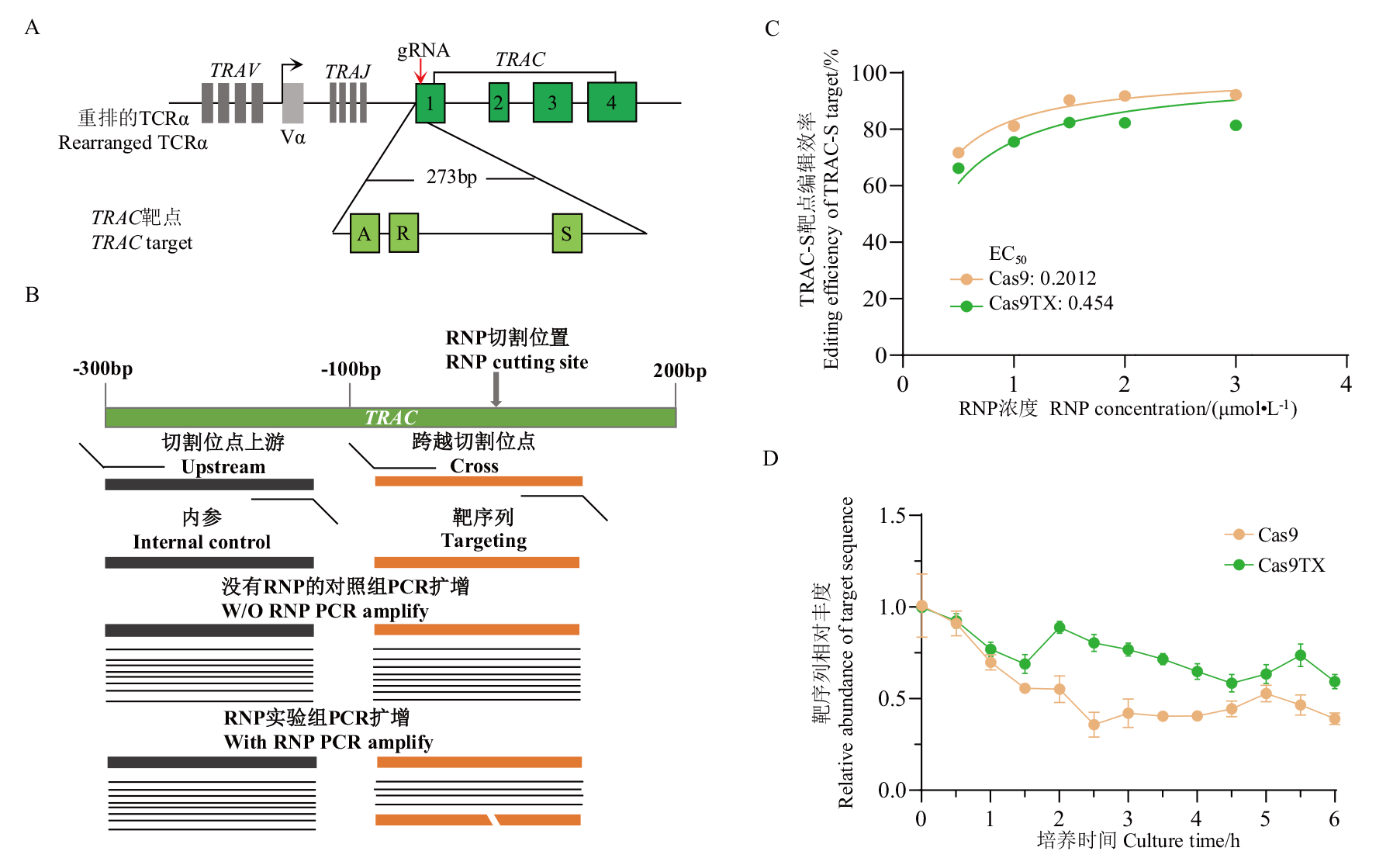
图3 T细胞内Cas9和Cas9TX的酶活性比较 A: TRAC gRNA靶点示意图;B: qPCR方案示意图;C:EC50 曲线;D: DSB生成曲线
Fig. 3 Comparison of enzyme activity of Cas9 and Cas9TX in T cell A: gRNA targeted sites. B: Scheme of qPCR strategy to measure unlinked DSBs. C: EC50 curve. D: Kinetics curve of DSB generation

图4 Cas9和Cas9TX在TRAC不同靶点的编辑效率 A:TIDE 分析Cas9在TRAC-S的敲除效率;B:TIDE分析Cas9TX 在TRAC-S的敲除效率;C:不同靶点敲除效率的统计结果
Fig. 4 Editing efficiencies of Cas9 and Cas9TX in different TRAC targets A: Knock out efficiency of Cas9 at TRAC-S site. B: Knock out efficiency of Cas9TX at TRAC-S site. C: Summary of knock out efficiencies at different targeted sites

图5 Cas9TX和Cas9TX RNP可降解供体模板
Fig. 5 DNA donor templates can be degraded by Cas9TX and Cas9TX RNP A: dsDNA; B: ssODN; C: GW dsDNA; D: 3' P ssODN. 1, donor template; 2, donor template with Cas9; 3, donor template with Cas9TX; 4, donor template with Cas9 RNP; 5, donor template with Cas9TX RNP
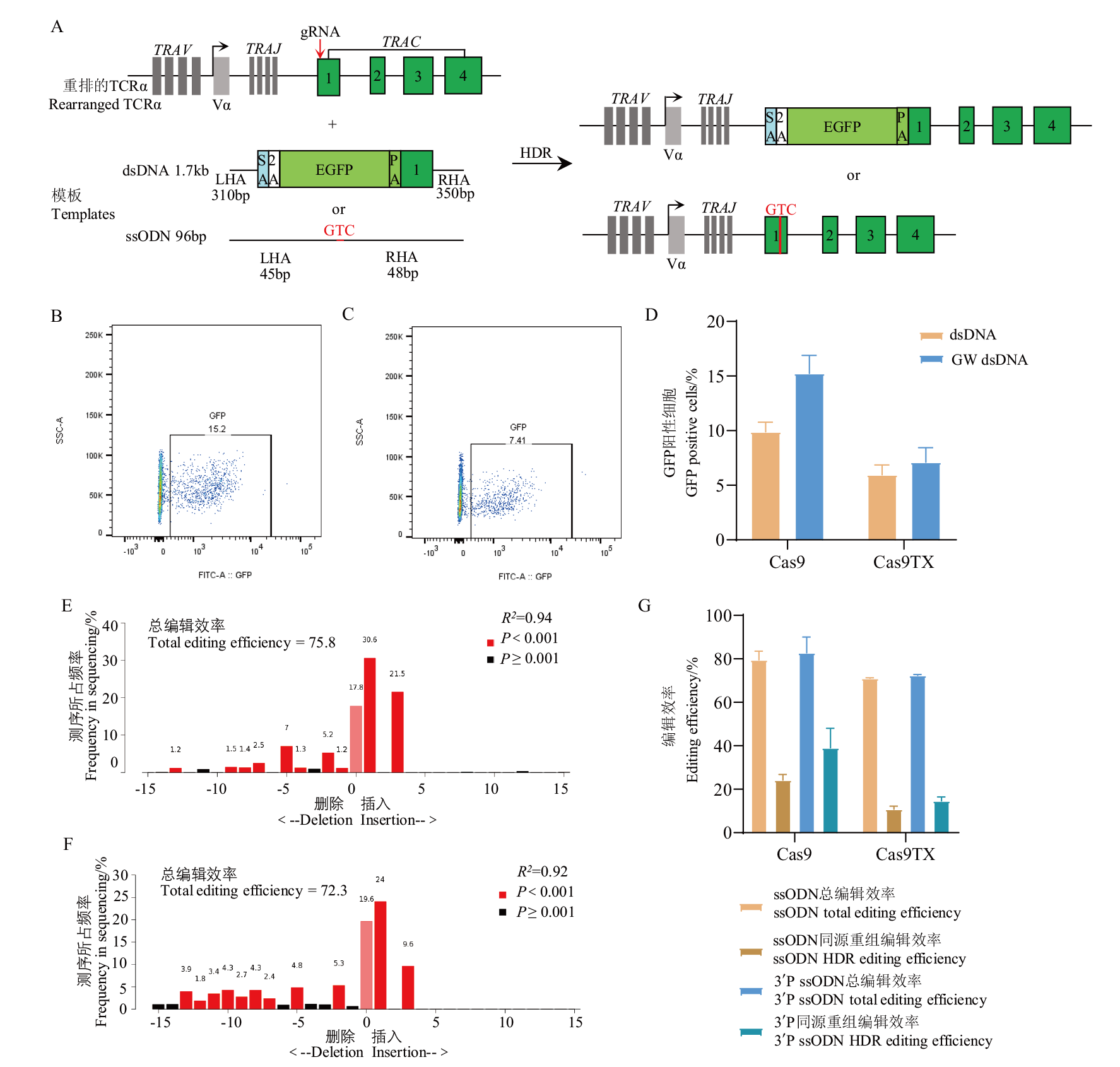
图6 供体模板稳定性修饰对定点敲入效率的影响 A:TRAC基因定点敲入方案模式图;B:流式分析Cas9组GFP表达比例;C:流式分析Cas9TX组GFP表达比例;D:GFP表达比例统计结果;E:TIDE分析Cas9组GTC碱基插入比例; F:TIDE分析Cas9TX组GTC碱基插入比例;G:GTC插入比例统计结果
Fig. 6 Effects of donor-template modification on the site-directed insertion efficiencies A: Scheme of 2A-GFP or GTC knock in strategies; B: flow cytometric analysis for GFP expression using Cas9; C: flow cytometric analysis for GFP expression using Cas9TX; D: statistical results of GFP knock in efficiencies; E: TIDE analysis for GTC knock in efficiencies using Cas9; F: TIDE analysis for GTC knock in efficiencies using Cas9TX;G: statistical results of GTC knock in efficiencies
| [1] |
Larson RC, Maus MV. Recent advances and discoveries in the mechanisms and functions of CAR T cells[J]. Nat Rev Cancer, 2021, 21(3): 145-161.
doi: 10.1038/s41568-020-00323-z pmid: 33483715 |
| [2] |
Russo-Carbolante EM, Picanço-Castro V, Alves DCC, et al. Integration pattern of HIV-1 based lentiviral vector carrying recombinant coagulation factor VIII in Sk-Hep and 293T cells[J]. Biotechnol Lett, 2011, 33(1): 23-31.
doi: 10.1007/s10529-010-0387-5 pmid: 20812025 |
| [3] |
Atianand MK, Fitzgerald KA. Molecular basis of DNA recognition in the immune system[J]. J Immunol, 2013, 190(5): 1911-1918.
doi: 10.4049/jimmunol.1203162 pmid: 23417527 |
| [4] |
Monjezi R, Miskey C, Gogishvili T, et al. Enhanced CAR T-cell engineering using non-viral Sleeping Beauty transposition from minicircle vectors[J]. Leukemia, 2017, 31(1): 186-194.
doi: 10.1038/leu.2016.180 pmid: 27491640 |
| [5] |
Foster JB, Choudhari N, Perazzelli J, et al. Purification of mRNA encoding chimeric antigen receptor is critical for generation of a robust T-cell response[J]. Hum Gene Ther, 2019, 30(2): 168-178.
doi: 10.1089/hum.2018.145 pmid: 30024272 |
| [6] |
Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements[J]. Nat Biotechnol, 2018, 36(8): 765-771.
doi: 10.1038/nbt.4192 pmid: 30010673 |
| [7] | Casini A, Olivieri M, Petris G, et al. A highly specific SpCas9 variant is identified by in vivo screening in yeast[J]. Nat Biotechnol, 2018, 36(3): 265-271. |
| [8] |
Chen BH, Gilbert LA, Cimini BA, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system[J]. Cell, 2013, 155(7): 1479-1491.
doi: 10.1016/j.cell.2013.12.001 pmid: 24360272 |
| [9] |
Yu Y, Guo YJ, Tian QQ, et al. An efficient gene knock-in strategy using 5'-modified double-stranded DNA donors with short homology arms[J]. Nat Chem Biol, 2020, 16(4): 387-390.
doi: 10.1038/s41589-019-0432-1 pmid: 31873222 |
| [10] |
Yin JH, Lu RS, Xin CC, et al. Cas9 exo-endonuclease eliminates chromosomal translocations during genome editing[J]. Nat Commun, 2022, 13(1): 1204.
doi: 10.1038/s41467-022-28900-w pmid: 35260581 |
| [11] |
Yin JH, Fang KL, Gao YX, et al. Safeguarding genome integrity during gene-editing therapy in a mouse model of age-related macular degeneration[J]. Nat Commun, 2022, 13(1): 7867.
doi: 10.1038/s41467-022-35640-4 pmid: 36550137 |
| [12] | Kath J, Du WJ, Pruene A, et al. Pharmacological interventions enhance virus-free generation of TRAC-replaced CAR Tcells[J]. Mol Ther Methods Clin Dev, 2022, 25: 311-330. |
| [13] |
Liu XJ, Zhang YP, Cheng C, et al. CRISPR-Cas9-mediated multiplex gene editing in CAR-T cells[J]. Cell Res, 2017, 27(1): 154-157.
doi: 10.1038/cr.2016.142 pmid: 27910851 |
| [14] | Yang M, Tkach D, Boyne A, et al. Optimized two-step electroporation process to achieve efficient nonviral-mediated gene insertion into primary T cells[J]. FEBS Open Bio, 2022, 12(1): 38-50. |
| [15] |
Harrigan JA, Fan JS, Momand J, et al. WRN exonuclease activity is blocked by DNA termini harboring 3' obstructive groups[J]. Mech Ageing Dev, 2007, 128(3): 259-266.
pmid: 17224176 |
| [16] |
Liao Q, Zheng JW, Wang B, et al. Efficient soluble expression and application of SpCas9 protein[J]. Food Science, 2023, 44(10): 150-157.
doi: 10.7506/spkx1002-6630-20220429-391 |
| [17] | 刘芳, 卢婷, 蔡梦迪, 等. Cas9蛋白的克隆表达、分离纯化及多克隆抗体制备[J]. 贵州医科大学学报, 2019, 44(7): 757-761, 766. |
| Liu F, Lu T, Cai MD, et al. Cloning expression, purification and polyclonal antibody preparation of Cas9 protein[J]. J Guizhou Med Univ, 2019, 44(7): 757-761, 766. | |
| [18] | Hu JZ, Yin JX. Fusion protein and use method thereof: WO2023011638(A1)[P]. 2023-02-09. |
| [1] | 侯文婷, 孙琳, 张艳军, 董合忠. 基因编辑技术在棉花种质创新和遗传改良中的应用[J]. 生物技术通报, 2024, 40(7): 68-77. |
| [2] | 朱恬仪, 孔桂美, 焦红梅, 郭停停, 乌日汗, 刘翠翠, 高成凤, 李国才. CRISPR/Cas9介导的adeG基因敲除大肠杆菌细菌模型的建立[J]. 生物技术通报, 2024, 40(2): 55-64. |
| [3] | 高登科, 马白荣, 郭怡莹, 刘薇, 刘田, 靳亚平, 江舟, 陈华涛. 利用CRISPR/Cas9技术构建Quaking敲除的小鼠胚胎成纤维细胞株[J]. 生物技术通报, 2024, 40(2): 65-72. |
| [4] | 张宏民, 龙雯, 劳筱清, 陈雯妍, 商雪梅, 王洪连, 王丽, 粟宏伟, 沈宏萍, 沈宏春. 利用CRISPR/Cas9技术构建Pmepa1基因敲除的TCMK1小鼠肾小管上皮细胞系[J]. 生物技术通报, 2024, 40(2): 73-79. |
| [5] | 陈小玲, 廖东庆, 黄尚飞, 陈英, 芦志龙, 陈东. 利用CRISPR/Cas9系统改造酿酒酵母的研究进展[J]. 生物技术通报, 2023, 39(8): 148-158. |
| [6] | 杨玉梅, 张坤晓. 应用CRISPR/Cas9技术建立ERK激酶相分离荧光探针定点整合的稳定细胞株[J]. 生物技术通报, 2023, 39(8): 159-164. |
| [7] | 施炜涛, 姚春鹏, 魏文康, 王蕾, 房元杰, 仝钰洁, 马晓姣, 蒋文, 张晓爱, 邵伟. 利用CRISPR/Cas9技术构建MDH2敲除细胞株及抗呕吐毒素效应研究[J]. 生物技术通报, 2023, 39(7): 307-315. |
| [8] | 刘晓燕, 祝振亮, 史广宇, 华梓宇, 杨晨, 张涌, 刘军. 乳腺生物反应器的表达优化策略[J]. 生物技术通报, 2023, 39(5): 77-91. |
| [9] | 程静雯, 曹磊, 张艳敏, 叶倩, 陈敏, 谭文松, 赵亮. CHO细胞多基因工程改造策略的建立及应用[J]. 生物技术通报, 2023, 39(2): 283-291. |
| [10] | 黄文莉, 李香香, 周炆婷, 罗莎, 姚维嘉, 马杰, 张芬, 沈钰森, 顾宏辉, 王建升, 孙勃. 利用CRISPR/Cas9技术靶向编辑青花菜BoZDS[J]. 生物技术通报, 2023, 39(2): 80-87. |
| [11] | 王兵, 赵会纳, 余婧, 陈杰, 骆梅, 雷波. 利用CRISPR/Cas9系统研究REVOLUTA参与烟草叶芽发育的调控[J]. 生物技术通报, 2023, 39(10): 197-208. |
| [12] | 李双喜, 华进联. 抗猪繁殖与呼吸障碍综合征基因编辑猪研究进展[J]. 生物技术通报, 2023, 39(10): 50-57. |
| [13] | 林蓉, 郑月萍, 徐雪珍, 李丹丹, 郑志富. 拟南芥ACOL8基因在乙烯合成与响应中的功能分析[J]. 生物技术通报, 2023, 39(1): 157-165. |
| [14] | 高伟欣, 黄火清, 赵晶, 张鑫, 杨宁, 杨浩萌. 应用于基因编辑的核糖核蛋白复合体的构建与活性验证[J]. 生物技术通报, 2022, 38(8): 60-68. |
| [15] | 刘静静, 刘晓蕊, 李琳, 王盈, 杨海元, 戴一凡. 利用CRISPR/Cas9技术建立OXTR基因敲除猪胎儿成纤维细胞系[J]. 生物技术通报, 2022, 38(6): 272-278. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||