








生物技术通报 ›› 2024, Vol. 40 ›› Issue (10): 262-274.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0408
梁婉凤1( ), 曾菁菁1, 胡若群1, 曹佳玉1, 郑涛2, 李銮2, 仇明月1, 梁晓英1, 陈莹1(
), 曾菁菁1, 胡若群1, 曹佳玉1, 郑涛2, 李銮2, 仇明月1, 梁晓英1, 陈莹1( )
)
收稿日期:2024-04-29
出版日期:2024-10-26
发布日期:2024-11-20
通讯作者:
陈莹,女,副教授,研究方向:观赏药用植物与园林生态;E-mail: 000q020057@fafu.edu.cn作者简介:梁婉凤,女,硕士研究生,研究方向:观赏药用植物与园林生态;E-mail: A576428098@163.com
基金资助:
LIANG Wan-feng1( ), ZENG Jing-jing1, HU Ruo-qun1, CAO Jia-yu1, ZHENG Tao2, LI Luan2, QIU Ming-yue1, LIANG Xiao-ying1, CHEN Ying1(
), ZENG Jing-jing1, HU Ruo-qun1, CAO Jia-yu1, ZHENG Tao2, LI Luan2, QIU Ming-yue1, LIANG Xiao-ying1, CHEN Ying1( )
)
Received:2024-04-29
Published:2024-10-26
Online:2024-11-20
摘要:
【目的】探究不同生长时期金线莲叶片中的类胡萝卜素的变化规律,解析其类胡萝卜素积累分子机制。【方法】选取不同生长时期(三月龄S3,六月龄S6,十月龄S10)金线莲叶片,采用液相色谱-质谱联用技术(LC-MS)和高通量转录组测序(RNA-seq)对不同生长时期的金线莲叶片进行分析。【结果】代谢组学结果表明参与叶片中类胡萝卜素合成的差异代谢物有23个,包括新黄质素、葡萄黄素、叶黄素和卡洛黄素等,转录组学结果表明差异代谢酶基因有21个,包括CYP97C1、CCD8、NCED、ZEP、PDS等,预测了ERF、MYB、C2H2、WRKY和bZIP等5种转录因子参与调控金线莲叶片类胡萝卜素的合成;并且进行了实时荧光定量PCR验证(RT-qPCR),实验表明选取的8个参与类胡萝卜素合成的酶基因在不同时期中的表达趋势与转录组学测序结果一致。【结论】金线莲不同生长时期ERF、MYB、C2H2等5种转录因子参与类胡萝卜素合成调控,与CYP97C1、CCD、NCED等5种关键差异酶基因关联明显,他们共同调控脱落酸醛、玉米黄质和叶黄素等6种关键差异代谢物。
梁婉凤, 曾菁菁, 胡若群, 曹佳玉, 郑涛, 李銮, 仇明月, 梁晓英, 陈莹. 转录组与代谢组分析不同生长时期金线莲类胡萝卜素的积累[J]. 生物技术通报, 2024, 40(10): 262-274.
LIANG Wan-feng, ZENG Jing-jing, HU Ruo-qun, CAO Jia-yu, ZHENG Tao, LI Luan, QIU Ming-yue, LIANG Xiao-ying, CHEN Ying. Transcriptional and Metabolomic Analysis of Carotenoid Accumulation in Anoectochilus roxburghii during Different Growth Periods[J]. Biotechnology Bulletin, 2024, 40(10): 262-274.

图1 不同生长阶段的金线莲 从左到右依次为3月龄、6月龄和10月龄。下同
Fig. 1 A. roxburghii at different growth stages 3-month(S3), 6-month(S6)and 10-month(S10)from left to right. The same below
| Gene | Gene ID | Primer sequence(5'-3') | Primer sequence(3'-5') |
|---|---|---|---|
| Actin | Reference gene | CTGTTTGCAGCCCAAGACAC | CAGCCTCCACAGCTTCAAGA |
| crtISO | TRINITY_DN4193_c0_g1 | TGGGAAGGACTTGACCGTCGTAG | ACCAACGCCACATCACACTTCTC |
| CYP97C1 | TRINITY_DN4271_c0_g1 | TCTCCATGATCCGACTGCAC | GACCGAAAGTCGAGAGTCTCA |
| DWARF27 | TRINITY_DN7678_c0_g1 | GCCTTGCTATGCCAACCTTT | TGCTGGGATTGGCCAAAGAA |
| crtB | TRINITY_DN28838_c0_g1 | TAGTGGGTGCAGGTTTCAGC | AATGGCTACATCTCCTGCCG |
| CCD8 | TRINITY_DN5697_c0_g1 | AGATTGCCGCACACCGAGTTG | AGCTCACCCTCCCATCGTTCTC |
| crtZ | TRINITY_DN67725_c0_g2 | AAGGCGCCAACACAGAATTG | CATGATTGCCGCCACCAAAT |
| ABA2 | TRINITY_DN8894_c0_g1 | GGTGTCACTGGCTCGAAAGT | GACACGCTCGCAAGAGAGAT |
| ZEP | TRINITY_DN9811_c0_g1 | TACCCGCACTTTAGGGAACG | CGCTAACACCAGTCCACCAA |
表1 RT-qPCR引物
Table 1 Primers for RT-qPCR
| Gene | Gene ID | Primer sequence(5'-3') | Primer sequence(3'-5') |
|---|---|---|---|
| Actin | Reference gene | CTGTTTGCAGCCCAAGACAC | CAGCCTCCACAGCTTCAAGA |
| crtISO | TRINITY_DN4193_c0_g1 | TGGGAAGGACTTGACCGTCGTAG | ACCAACGCCACATCACACTTCTC |
| CYP97C1 | TRINITY_DN4271_c0_g1 | TCTCCATGATCCGACTGCAC | GACCGAAAGTCGAGAGTCTCA |
| DWARF27 | TRINITY_DN7678_c0_g1 | GCCTTGCTATGCCAACCTTT | TGCTGGGATTGGCCAAAGAA |
| crtB | TRINITY_DN28838_c0_g1 | TAGTGGGTGCAGGTTTCAGC | AATGGCTACATCTCCTGCCG |
| CCD8 | TRINITY_DN5697_c0_g1 | AGATTGCCGCACACCGAGTTG | AGCTCACCCTCCCATCGTTCTC |
| crtZ | TRINITY_DN67725_c0_g2 | AAGGCGCCAACACAGAATTG | CATGATTGCCGCCACCAAAT |
| ABA2 | TRINITY_DN8894_c0_g1 | GGTGTCACTGGCTCGAAAGT | GACACGCTCGCAAGAGAGAT |
| ZEP | TRINITY_DN9811_c0_g1 | TACCCGCACTTTAGGGAACG | CGCTAACACCAGTCCACCAA |
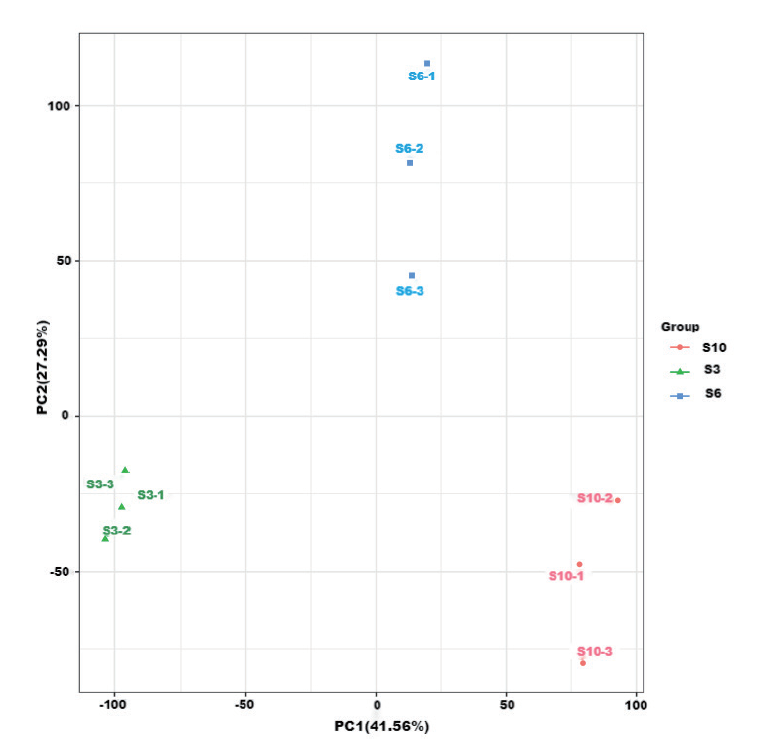
图2 样本主成分分析图 图中同一颜色为一个组别,数字标号指同组的不同重复,如S3-3是3月龄组别的第3个重复,下同
Fig. 2 Principal component analysis plot for samples The same color in the figure is a group, and the numerical labels refer to different replicates of the same group, e.g., S3-3 is the third replicate of the 3-month group, the same below
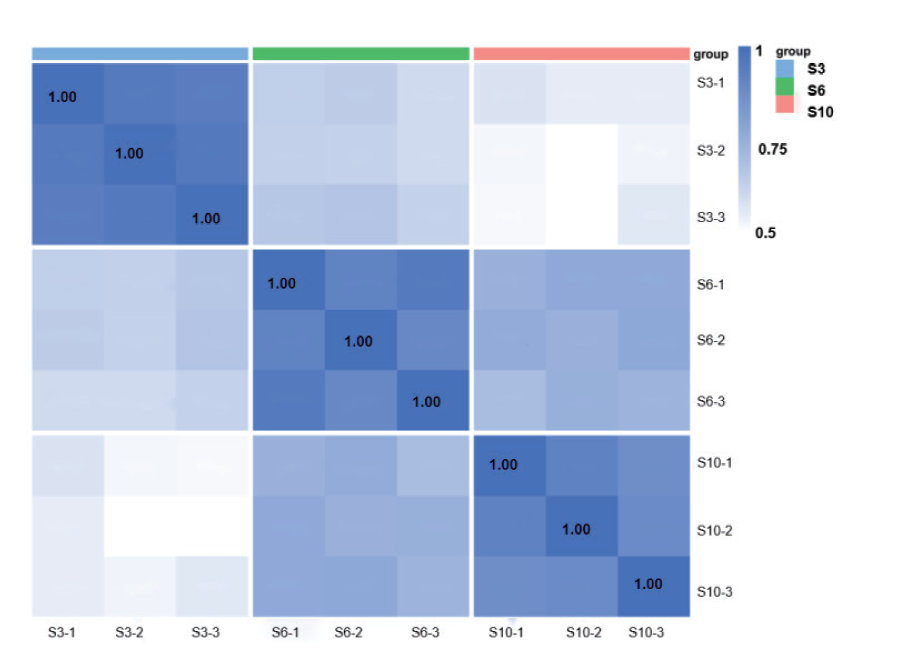
图3 样本相关性热图 图上蓝色、绿色、粉色代表的是分别是3、6、10月龄,框内蓝色颜色越深相关性越深
Fig. 3 Heat map of sample correlation The blue, green, and pink colors on the chart represent 3-, 6-, and 10-month, respectively; the darker the blue color in the box, the deeper the correlation
| 代谢物 Metabolite | S3_vs_S6 regulated | S6_vs_S10 regulated | S3_vs_S10 regulated | 分子式Formula | KEGG注释 KEGG annotation |
|---|---|---|---|---|---|
| 1'-羟基-gamma-胡萝卜素葡萄糖苷 1'-Hydroxy-gamma-carotene glucoside | Unchanged | Up | Unchanged | C46H68O6 | C15861 |
| 脱落酸葡萄糖酯Abscisic acid glucose ester | Up | Unchanged | Unchanged | C21H30O9 | C15970 |
| 酮基R.g.-Keto III | Up | Unchanged | Up | C42H60O4 | C16278 |
| 辣椒素Capsorubin | Unchanged | Unchanged | Down | C40H56O4 | C08585 |
| 羟基螺旋黄质Hydroxyspirilloxanthin | Down | Unchanged | Down | C41H58O2 | C15879 |
| 新黄质素Neoxanthin | Up | Unchanged | Up | C40H56O4 | C08606 |
| 葡萄黄素Staphyloxanthin | Unchanged | Up | Up | C51H78O8 | C16148 |
| 诺吐黄素Nostoxanthin | Unchanged | Up | Up | C40H56O4 | C16284 |
| 2-酮基螺旋体黄质2-Ketospirilloxanthin | Unchanged | Unchanged | Down | C42H58O3 | C15884 |
| 羟基氯巴坦Hydroxychlorobactene | Down | Up | Unchanged | C40H54O | C15911 |
| 3-羟基叶茶烯酮3-Hydroxyechinenone | Unchanged | Up | Unchanged | C40H54O2 | C15966 |
| 三苯甲内酯 ABC 环Strigolactone ABC-rings | Down | Down | Down | C14H18O3 | C18036 |
| 8'-羟基琥珀酸酯8'-Hydroxyabscisate | Unchanged | Up | Up | C15H20O5 | C15514 |
| 脱落酸醛Abscisic aldehyde | Down | Unchanged | Down | C15H20O3 | C13455 |
| 羟基氯巴坦葡萄糖苷Hydroxychlorobactene glucoside | Down | Unchanged | Down | C46H64O6 | C15914 |
| (2'S)-脱氧木酚 2'-alpha-L-岩藻糖苷 (2'S)-Deoxymyxol 2'-alpha-L-fucoside | Down | Unchanged | Down | C46H66O6 | C15940 |
| (2'S)-脱氧木酚 2'-(2,4-二-O-甲基-alpha-L-岩藻糖苷) (2'S)-Deoxymyxol 2'-(2,4-di-O-methyl-alpha-L-fucoside) | Up | Down | Unchanged | C48H70O6 | C15935 |
| (3R,2'S)-2'-(2,4-二-O-甲基-alpha-L-岩藻糖苷)岩藻醇 (3R,2'S)-Myxol 2'-(2,4-di-O-methyl-alpha-L-fucoside) | Down | Up | Unchanged | C48H70O7 | C15937 |
| 腓尼基黄质Phoenicoxanthin | Up | Unchanged | Up | C40H52O3 | C15967 |
| 玉米黄质Zeinoxanthin | Unchanged | Up | Unchanged | C40H56O | C08590 |
| 腺嘌呤Adonixanthin | Up | Unchanged | Up | C40H54O3 | C15968 |
| 叶黄素Lutein | Up | Unchanged | Up | C40H56O2 | C08601 |
| 卡洛黄素Caloxanthin | Up | Unchanged | Unchanged | C40H56O3 | C16282 |
表2 不同生长时期金线莲叶片中差异类胡萝卜素代谢物
Table 2 Differential carotenoid metabolites in the leaves of A. roxburghii at different growth stages
| 代谢物 Metabolite | S3_vs_S6 regulated | S6_vs_S10 regulated | S3_vs_S10 regulated | 分子式Formula | KEGG注释 KEGG annotation |
|---|---|---|---|---|---|
| 1'-羟基-gamma-胡萝卜素葡萄糖苷 1'-Hydroxy-gamma-carotene glucoside | Unchanged | Up | Unchanged | C46H68O6 | C15861 |
| 脱落酸葡萄糖酯Abscisic acid glucose ester | Up | Unchanged | Unchanged | C21H30O9 | C15970 |
| 酮基R.g.-Keto III | Up | Unchanged | Up | C42H60O4 | C16278 |
| 辣椒素Capsorubin | Unchanged | Unchanged | Down | C40H56O4 | C08585 |
| 羟基螺旋黄质Hydroxyspirilloxanthin | Down | Unchanged | Down | C41H58O2 | C15879 |
| 新黄质素Neoxanthin | Up | Unchanged | Up | C40H56O4 | C08606 |
| 葡萄黄素Staphyloxanthin | Unchanged | Up | Up | C51H78O8 | C16148 |
| 诺吐黄素Nostoxanthin | Unchanged | Up | Up | C40H56O4 | C16284 |
| 2-酮基螺旋体黄质2-Ketospirilloxanthin | Unchanged | Unchanged | Down | C42H58O3 | C15884 |
| 羟基氯巴坦Hydroxychlorobactene | Down | Up | Unchanged | C40H54O | C15911 |
| 3-羟基叶茶烯酮3-Hydroxyechinenone | Unchanged | Up | Unchanged | C40H54O2 | C15966 |
| 三苯甲内酯 ABC 环Strigolactone ABC-rings | Down | Down | Down | C14H18O3 | C18036 |
| 8'-羟基琥珀酸酯8'-Hydroxyabscisate | Unchanged | Up | Up | C15H20O5 | C15514 |
| 脱落酸醛Abscisic aldehyde | Down | Unchanged | Down | C15H20O3 | C13455 |
| 羟基氯巴坦葡萄糖苷Hydroxychlorobactene glucoside | Down | Unchanged | Down | C46H64O6 | C15914 |
| (2'S)-脱氧木酚 2'-alpha-L-岩藻糖苷 (2'S)-Deoxymyxol 2'-alpha-L-fucoside | Down | Unchanged | Down | C46H66O6 | C15940 |
| (2'S)-脱氧木酚 2'-(2,4-二-O-甲基-alpha-L-岩藻糖苷) (2'S)-Deoxymyxol 2'-(2,4-di-O-methyl-alpha-L-fucoside) | Up | Down | Unchanged | C48H70O6 | C15935 |
| (3R,2'S)-2'-(2,4-二-O-甲基-alpha-L-岩藻糖苷)岩藻醇 (3R,2'S)-Myxol 2'-(2,4-di-O-methyl-alpha-L-fucoside) | Down | Up | Unchanged | C48H70O7 | C15937 |
| 腓尼基黄质Phoenicoxanthin | Up | Unchanged | Up | C40H52O3 | C15967 |
| 玉米黄质Zeinoxanthin | Unchanged | Up | Unchanged | C40H56O | C08590 |
| 腺嘌呤Adonixanthin | Up | Unchanged | Up | C40H54O3 | C15968 |
| 叶黄素Lutein | Up | Unchanged | Up | C40H56O2 | C08601 |
| 卡洛黄素Caloxanthin | Up | Unchanged | Unchanged | C40H56O3 | C16282 |

图4 不同生长时期金线莲叶片差异类胡萝卜素物质代谢物聚类热图
Fig. 4 Clustering heat map of metabolites of differential carotenoid substances in the leaves of A. roxburghii at different growth stages
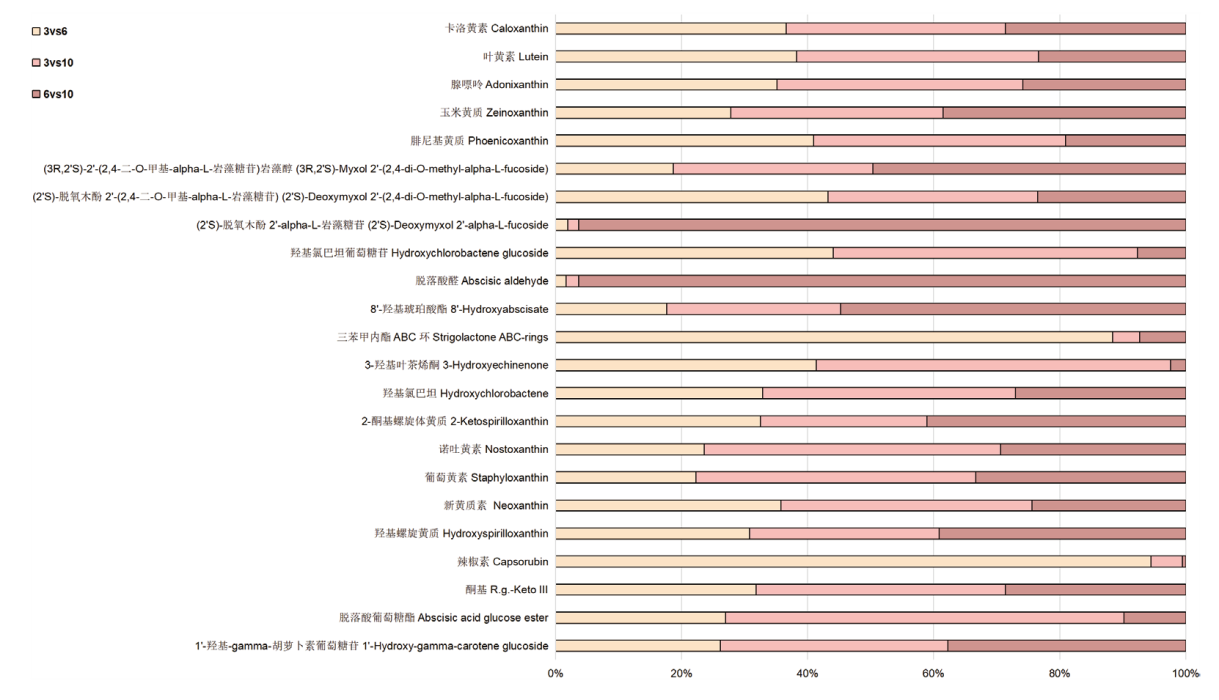
图5 不同生长时期金线莲叶片差异类胡萝卜素物质代谢物变化比例图 浅黄色柱状代表3月龄和6月龄的比较,粉色柱状代表3月龄和10月龄的比较,棕色柱状代表6月龄和10月龄的比较,柱状越长变化越大,框内数字代表变化倍数
Fig. 5 Proportion of changes in the metabolites of different carotenoids in the leaves of A. roxburghii at different growth stages Light yellow bars indicate the comparison between 3-month and 6-month, pink bars indicate the comparison between 3-month and 10-month, and brown bars indicate the comparison between 6-month and 10-month. The longer the bar, the greater the change, and the numbers in the box indicate multiples of the change
| 基因 Gene | ID | S3 VS S6 DESeq regulated | S6 VS S10 DESeq regulated | S3 VS S10 DESeq regulated | 对应酶 Corresponding enzyme | 酶KO Enzyme KO |
|---|---|---|---|---|---|---|
| CCD8 | TRINITY_DN1716_c0_g1 | Down | Normal | Normal | [EC:1.13.11.68] | K17912 |
| TRINITY_DN5697_c0_g1 | Down | Normal | Down | [EC:1.13.11.69 1.13.11.70] | K17913 | |
| TRINITY_DN8190_c0_g1 | Down | Normal | Down | [EC:1.13.11.69 1.13.11.70] | K02293 | |
| PDS | TRINITY_DN9_c1_g1 | Normal | Normal | Up | [EC:1.3.5.5] | K02293 |
| NCED | TRINITY_DN11998_c0_g1 | Up | Normal | Up | [EC:1.13.11.51] | K09840 |
| TRINITY_DN9005_c0_g1 | Up | Up | Up | [EC:1.13.11.51] | K15746 | |
| crtISO | TRINITY_DN4193_c0_g1 | Up | Normal | Normal | [EC:5.2.1.13] | K09835 |
| TRINITY_DN1162_c2_g1 | Down | Normal | Normal | [EC:5.2.1.13] | K09843 | |
| crtZ | TRINITY_DN6176_c0_g1 | Up | Normal | Normal | [EC:1.14.15.24] | K15746 |
| TRINITY_DN67725_c0_g2 | Down | Up | Normal | [EC:1.14.15.24] | K09838 | |
| crtB | TRINITY_DN28838_c0_g1 | Up | Up | Normal | [EC:2.5.1.32] | K02291 |
| TRINITY_DN4528_c0_g1 | Down | Down | Normal | [EC:2.5.1.32] | K09841 | |
| CYP97C1 | TRINITY_DN5659_c0_g1 | Down | Normal | Down | [EC:1.14.14.137] | K09843 |
| TRINITY_DN4271_c0_g1 | Down | Normal | Down | [EC:1.14.14.137] | K14595 | |
| TRINITY_DN586_c0_g1 | Down | Down | Normal | [EC:1.14.14.158] | K09837 | |
| ZEP | TRINITY_DN9811_c0_g1 | Up | Normal | Normal | [EC:1.14.15.21] | K09838 |
| LUT5, CYP97A3 | TRINITY_DN2395_c0_g1 | Up | Normal | Normal | [EC:1.14.15.21] | K02293 |
| DWARF27 | TRINITY_DN7678_c0_g1 | Up | Up | Normal | [EC:5.2.1.14] | K17911 |
| ABA2 | TRINITY_DN8894_c0_g1 | Down | Normal | Normal | [EC:1.1.1.28] | K09841 |
| crtL2 | TRINITY_DN7101_c0_g1 | Up | Normal | Up | [EC:5.5.1.18] | K06444 |
| AOG | TRINITY_DN13229_c0_g1 | Down | Down | Normal | [EC:2.4.1.26] | K14595 |
表3 不同生长时期金线莲叶片中差异类胡萝卜素酶基因
Table 3 Differential carotenoid enzyme genes in A. roxburghii leaves at different growth stages
| 基因 Gene | ID | S3 VS S6 DESeq regulated | S6 VS S10 DESeq regulated | S3 VS S10 DESeq regulated | 对应酶 Corresponding enzyme | 酶KO Enzyme KO |
|---|---|---|---|---|---|---|
| CCD8 | TRINITY_DN1716_c0_g1 | Down | Normal | Normal | [EC:1.13.11.68] | K17912 |
| TRINITY_DN5697_c0_g1 | Down | Normal | Down | [EC:1.13.11.69 1.13.11.70] | K17913 | |
| TRINITY_DN8190_c0_g1 | Down | Normal | Down | [EC:1.13.11.69 1.13.11.70] | K02293 | |
| PDS | TRINITY_DN9_c1_g1 | Normal | Normal | Up | [EC:1.3.5.5] | K02293 |
| NCED | TRINITY_DN11998_c0_g1 | Up | Normal | Up | [EC:1.13.11.51] | K09840 |
| TRINITY_DN9005_c0_g1 | Up | Up | Up | [EC:1.13.11.51] | K15746 | |
| crtISO | TRINITY_DN4193_c0_g1 | Up | Normal | Normal | [EC:5.2.1.13] | K09835 |
| TRINITY_DN1162_c2_g1 | Down | Normal | Normal | [EC:5.2.1.13] | K09843 | |
| crtZ | TRINITY_DN6176_c0_g1 | Up | Normal | Normal | [EC:1.14.15.24] | K15746 |
| TRINITY_DN67725_c0_g2 | Down | Up | Normal | [EC:1.14.15.24] | K09838 | |
| crtB | TRINITY_DN28838_c0_g1 | Up | Up | Normal | [EC:2.5.1.32] | K02291 |
| TRINITY_DN4528_c0_g1 | Down | Down | Normal | [EC:2.5.1.32] | K09841 | |
| CYP97C1 | TRINITY_DN5659_c0_g1 | Down | Normal | Down | [EC:1.14.14.137] | K09843 |
| TRINITY_DN4271_c0_g1 | Down | Normal | Down | [EC:1.14.14.137] | K14595 | |
| TRINITY_DN586_c0_g1 | Down | Down | Normal | [EC:1.14.14.158] | K09837 | |
| ZEP | TRINITY_DN9811_c0_g1 | Up | Normal | Normal | [EC:1.14.15.21] | K09838 |
| LUT5, CYP97A3 | TRINITY_DN2395_c0_g1 | Up | Normal | Normal | [EC:1.14.15.21] | K02293 |
| DWARF27 | TRINITY_DN7678_c0_g1 | Up | Up | Normal | [EC:5.2.1.14] | K17911 |
| ABA2 | TRINITY_DN8894_c0_g1 | Down | Normal | Normal | [EC:1.1.1.28] | K09841 |
| crtL2 | TRINITY_DN7101_c0_g1 | Up | Normal | Up | [EC:5.5.1.18] | K06444 |
| AOG | TRINITY_DN13229_c0_g1 | Down | Down | Normal | [EC:2.4.1.26] | K14595 |
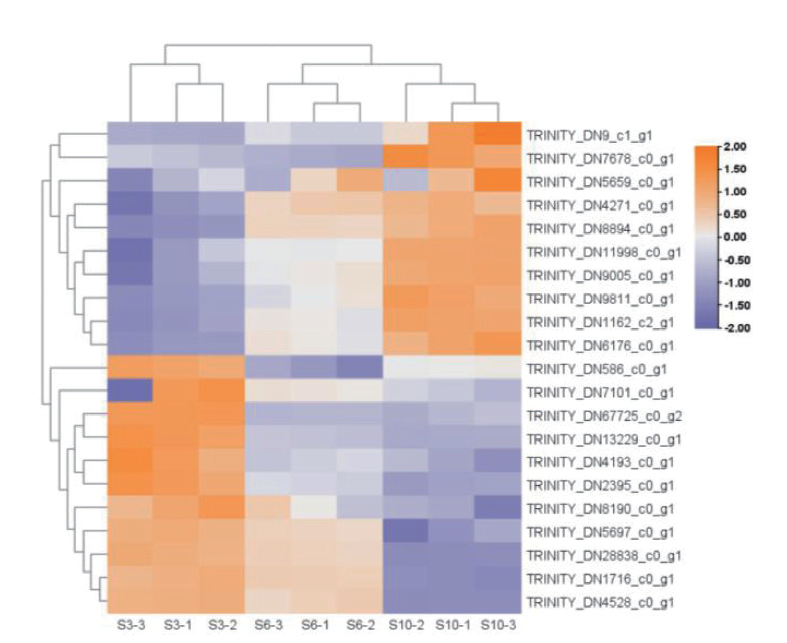
图6 类胡萝卜素合成相关差异基因聚类热图 热图中橙色代表高表达水平紫色代表低表达水平,颜色越深程度越深
Fig. 6 Heat map of carotenoid synthesis-related differential gene clustering In the heat map, orange indicates high expression level and purple indicates low expression level. The darker the color, the deeper the degree
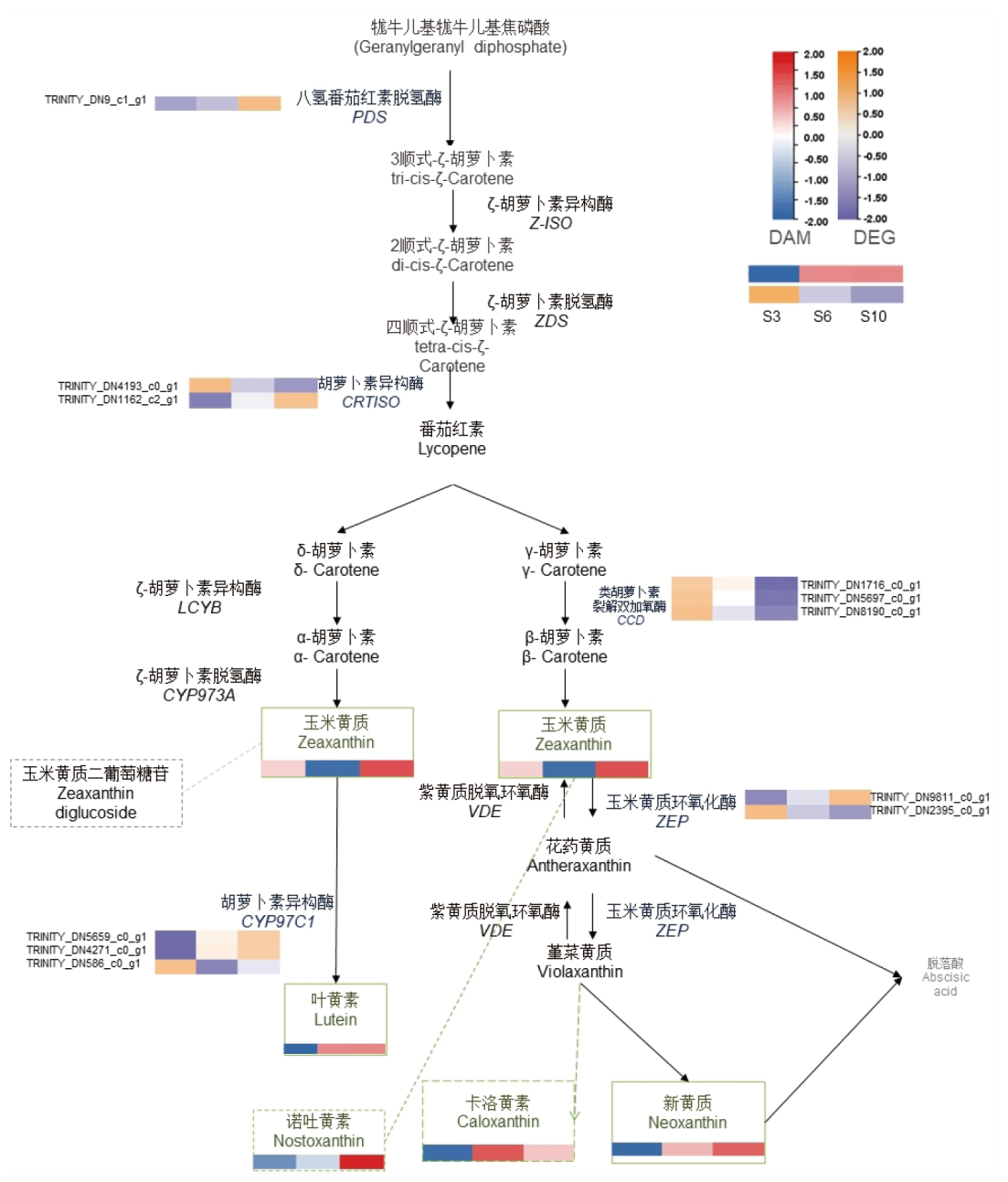
图7 类胡萝卜素转录代谢联合分析图 图中实线框和虚线框分别代表主要通路及非主要,橙色英文和绿色中文为差异基因和差异代谢物,其中红色蓝色代表着差异代谢物在热图中表达由高到低,而橙色紫色则代表着差异转录基因在热图中表达由高到低
Fig. 7 Co-analysis map of carotenoid transcript metabolism Solid and dashed boxes indicate major pathways and non-major, respectively; orange English and green Chinese are differential genes and differential metabolites, where red-blue indicates differential metabolites expressed from high to low in the heat map, and orange-purple indicates differential transcribed genes expressed from high to low in the heat map

图8 类胡萝卜素转录代谢中相关转录因子与酶基因调控关系图 绿色图形代表酶基因,紫色图形代表转录因子,实线和虚线分别代表正负相关性,线条越粗相关性越强
Fig. 8 Regulation correlation between transcription factors and enzyme genes in carotenoid transcription and metabolism Green graph indicates enzyme genes, purple graph indicates transcription factors, solid and dashed lines indicate positive and negative correlations, respectively; the thicker the line, the stronger the correlation
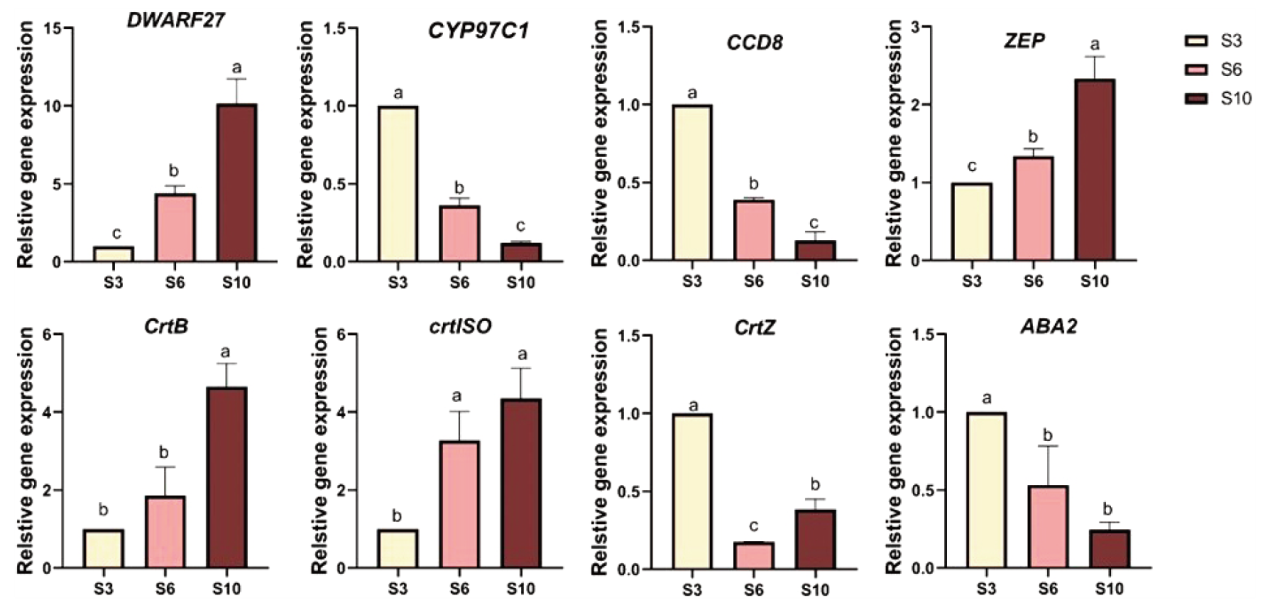
图9 RT-qPCR 分析不同生长时期金线莲叶片中的差异酶基因 浅黄色柱状和S3代表3月龄,粉色柱状和S6代表6月龄,棕色柱状和S10代表10月龄。不同字母表示在0.05水平上差异性显著(P<0.05)
Fig. 9 RT-qPCR analysis of different enzymes genes in A. roxburghii leaves at different growth stages Light yellow bars and S3 indicate 3 months of age, pink bars and S6 indicate 6 months of age, and brown bars and S10 indicate 10 months of age. Different letters indicate significant differences at the 0.05 level(P < 0.05)
| [1] | 郑丽香. 金线莲的资源调查及生药学研究[D]. 福州: 福建中医药大学, 2018. |
| Zheng LX. Resource investigation and pharmacognosy study of anoectochilus roxburghii[D]. Fuzhou: Fujian University of Traditional Chinese Medicine, 2018. | |
| [2] | 李荣峰. 金线莲研究进展[J]. 安徽农学通报, 2018, 24(24): 27-29. |
| Li RF. The research progress about Anoectochilus roxburghii[J]. Anhui Agric Sci Bull, 2018, 24(24): 27-29. | |
| [3] | Miki W. Biological functions and activities of animal carotenoids[J]. Pure Appl Chem, 1991, 63(1): 141-146. |
| [4] | 董彩英. 辣椒果实类胡萝卜素的积累特点及其与果色的关系研究[D]. 扬州: 扬州大学, 2009. |
| Dong CY. Studies on the accumulation characteristics of carotenoids in pepper fruits and their relationship with fruit color[D]. Yangzhou: Yangzhou University, 2009. | |
| [5] |
Nagao A. Absorption and function of dietary carotenoids[J]. Forum Nutr, 2009, 61: 55-63.
doi: 10.1159/000212738 pmid: 19367110 |
| [6] |
Fraser P. The biosynthesis and nutritional uses of carotenoids[J]. Prog Lipid Res, 2004, 43(3): 228-265.
doi: 10.1016/j.plipres.2003.10.002 pmid: 15003396 |
| [7] |
Nishino H, Murakoshi M, et al. Cancer prevention by carotenoids[J]. Arch Biochem Biophys, 2009, 483(2): 165-168.
doi: 10.1016/j.abb.2008.09.011 pmid: 18848517 |
| [8] | 陶俊, 张上隆, 等. 类胡萝卜素合成的相关基因及其基因工程[J]. 生物工程学报, 2002, 18(3): 276-281. |
| Tao J, Zhang SL, et al. Gene and gene engineering of carotenoid biosynthesis[J]. Chin J Biotechnol, 2002, 18(3): 276-281. | |
| [9] |
Ito M, Yamano Y, et al. Carotenoid synthesis: retrospect and recent progress[J]. Arch Biochem Biophys, 2009, 483(2): 224-228.
doi: 10.1016/j.abb.2008.11.021 pmid: 19068206 |
| [10] |
赵艳侠, 张晶莹, 等. ‘重瓣红’玫瑰不同花发育阶段转录和代谢差异分析[J]. 生物技术通报, 2023, 39(3): 184-195.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-0729 |
| Zhao YX, Zhang JY, et al. Analyses of transcription and metabolic differential in the flower development processes of ‘Rose rugosa cv.Plena’[J]. Biotechnol Bull, 2023, 39(3): 184-195. | |
| [11] | 杨程, 刘小玉, 周丽霞, 等. 油棕外果皮类胡萝卜素合成的转录代谢联合分析[J/OL]. 分子植物育种, 2024: 1-16. |
| Yang C, Liu XY, Zhou LX, et al. Joint transcriptional-metabolic analysis of carotenoid synthesis in oil palm exocarp[J/OL]. Molecular Plant Breeding, 2024: 1-16. | |
| [12] | 高雪倩, 贾云彭, 李昕悦, 等. 建兰不同花器官花香代谢差异的转录组分析[J/OL]. 分子植物育种, 2024: 1-11. |
| Gao XQ, Jia YP, Li XY, et al. Transcriptome analysis of differences in floral flavor metabolism of different floral organs in Jianlan[J/OL]. Molecular Plant Breeding, 2024: 1-11. | |
| [13] | 王露凡, 杨晓涵, 等. 桃果实冷害形成过程关键代谢途径的转录组学研究[J]. 食品与生物技术学报, 2024, 43(3): 66-75. |
| Wang LF, Yang XH, et al. Transcriptomics of key metabolic pathways during cold damage formation in peach fruit[J]. Journal of Food Science and Biotechnology, 2024, 43(3): 66-75. | |
| [14] | 但英, 陈若, 李翠新, 等. 组培福建金线莲生物活性成分提升分析[J/OL]. 分子植物育种, 2023:1-19. |
| Dan Y, Chen R, Li CX, et al. Analysis on the lmprovement of bioactive constituents in tissue culture of Anoectochilus roxburghi[J/OL]. Molecular Plant Breeding, 2023:1-19. | |
| [15] | 周媛媛, 赵目聪, 陈蕾, 等. 金线莲降血糖作用及作用机制研究进展[J]. 广州化工, 2023, 51(13): 21-24. |
| Zhou YY, Zhao MC, Chen L, et al. Research progress on hypoglycemic effect and mechanism of Anoectochilus roxburghii[J]. Guangzhou Chem Ind, 2023, 51(13): 21-24. | |
| [16] | 邵玲, 关玉媛, 郭林洁, 等. DA-6与椰汁对金线莲组培苗壮苗生根的影响[J]. 中药材, 2023, 46(7): 1603-1607. |
| Shao L, Guan YY, Guo LJ, et al. Effects of DA-6 and coconut juice on rooting of tissue culture seedlings of Anoectochilus roxburghii[J]. J Chin Med Mater, 2023, 46(7): 1603-1607. | |
| [17] | Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2[J]. Genome Biol, 2014, 15(12): 550. |
| [18] | Han YJ, Wang XH, Chen WC, et al. Differential expression of carotenoid-related genes determines diversified carotenoid coloration in flower petal of Osmanthus fragrans[J]. Tree Genet Genomes, 2014, 10(2): 329-338. |
| [19] | Wang YG, Zhang C, Dong B, et al. Carotenoid accumulation and its contribution to flower coloration of Osmanthus fragrans[J]. Front Plant Sci, 2018, 9: 1499. |
| [20] |
应震, 张晶, 殷恒福, 等. 茶花红叶芽变品种‘金华美女’叶色突变相关主要化学成分含量变化[J]. 园艺学报, 2017, 44(4): 723-732.
doi: 10.16420/j.issn.0513-353x.2016-0714 |
| Ying Z, Zhang J, Yin HF, et al. Content change of main chemical component relating to the red leaf of bud mutation camellia variety‘Jinhua Meinu’[J]. Acta Horticulturae Sinica, 2017, 44(4): 723-732. | |
| [21] | 王贵一, 孟嘉珺, 许文静, 等. 不同品种芒果的营养成分及风味物质分析[J]. 食品工业科技, 2022, 43(1): 71-79. |
| Wang GY, Meng JJ, Xu WJ, et al. Analysis of nutritional components and flavor substances of different varieties of mangoes[J]. Sci Technol Food Ind, 2022, 43(1): 71-79. | |
| [22] | Yu YG, Chen XP, Zheng Q. Metabolomic profiling of carotenoid constituents in Physalis peruviana during different growth stages by LC-MS/MS technology[J]. J Food Sci, 2019, 84(12): 3608-3613. |
| [23] |
Huang H, Lu CF, Ma S, et al. Different colored Chrysanthemum × morifolium cultivars represent distinct plastid transformation and carotenoid deposit patterns[J]. Protoplasma, 2019, 256(6): 1629-1645.
doi: 10.1007/s00709-019-01406-x pmid: 31267226 |
| [24] |
Aharoni A, De Vos CH, et al. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco[J]. Plant J, 2001, 28(3): 319-332.
doi: 10.1046/j.1365-313x.2001.01154.x pmid: 11722774 |
| [25] | 房强. 香雪兰类胡萝卜素裂解双加氧酶(FhCCDs)基因克隆与功能鉴定[D]. 长春: 东北师范大学, 2020. |
| Fang Q. Cloning and functional identification of carotenoid-cleaving dioxygenase(FhCCDs)gene from Ceylon[D]. Changchun: Northeast Normal University, 2020. | |
| [26] | Yao YX, Jia L, Cheng Y, et al. Evolutionary origin of the carotenoid cleavage oxygenase family in plants and expression of pepper genes in response to abiotic stresses[J]. Front Plant Sci, 2022, 12: 792832. |
| [27] |
Diao QN, Tian SB, Cao YY, et al. Transcriptome analysis reveals association of carotenoid metabolism pathway with fruit color in melon[J]. Sci Rep, 2023, 13(1): 5004.
doi: 10.1038/s41598-023-31432-y pmid: 36973323 |
| [28] | Arango J, Jourdan M, Geoffriau E, et al. Carotene hydroxylase activity determines the levels of both α-carotene and total carotenoids in orange carrots[J]. Plant Cell, 2014, 26(5): 2223-2233. |
| [29] |
Gonzalez-Jorge S, Mehrshahi P, et al. ZEAXANTHIN EPOXIDASE activity potentiates carotenoid degradation in maturing seed[J]. Plant Physiol, 2016, 171(3): 1837-1851.
doi: 10.1104/pp.16.00604 pmid: 27208224 |
| [30] | Ma XW, Zheng B, et al. Carotenoid accumulation and expression of carotenoid biosynthesis genes in mango flesh during fruit development and ripening[J]. Sci Hortic, 2018, 237: 201-206. |
| [31] | Liu HY, Mao JH, Yan SJ, et al. Evaluation of carotenoid biosynthesis, accumulation and antioxidant activities in sweetcorn(Zea mays L.) during kernel development[J]. Int J Food Sci Tech, 2018, 53(2): 381-388. |
| [32] | Liang MH, Li XY. Involvement of transcription factors and regulatory proteins in the regulation of carotenoid accumulation in plants and algae[J]. J Agric Food Chem, 2023, 71(48): 18660-18673. |
| [33] | He Y, Li MR, Wang YH, et al. The R2R3-MYB transcription factor MYB44 modulates carotenoid biosynthesis in Ulva prolifera[J]. Algal Res, 2022, 62: 102578. |
| [34] | Owji H, Hajiebrahimi A, Seradj H, et al. Identification and functional prediction of stress responsive AP2/ERF transcription factors in Brassica napus by genome-wide analysis[J]. Comput Biol Chem, 2017, 71: 32-56. |
| [35] |
Wang XB, Zeng WF, Ding YF, et al. Peach ethylene response factor PpeERF2 represses the expression of ABA biosynthesis and cell wall degradation genes during fruit ripening[J]. Plant Sci, 2019, 283: 116-126.
doi: S0168-9452(18)31012-4 pmid: 31128681 |
| [36] | Qi XN, Xiao YY, Fan ZQ, et al. A banana fruit transcriptional repressor MaERF10 interacts with MaJAZ3 to strengthen the repression of JA biosynthetic genes involved in MeJA-mediated cold tolerance[J]. Postharvest Biol Technol, 2016, 120: 222-231. |
| [37] |
王佳慧, 顾凯迪, 王楚堃, 等. 苹果乙烯响应因子MdERF72对非生物胁迫的响应[J]. 中国农业科学, 2019, 52(23): 4374-4385.
doi: 10.3864/j.issn.0578-1752.2019.23.017 |
| Wang JH, Gu KD, Wang CK, et al. Analysis of apple ethylene response factor MdERF72 to abiotic stresses[J]. Sci Agric Sin, 2019, 52(23): 4374-4385. | |
| [38] |
Zhang J, Yin XR, Li H, et al. ETHYLENE RESPONSE FACTOR39-MYB8 complex regulates low-temperature-induced lignification of loquat fruit[J]. J Exp Bot, 2020, 71(10): 3172-3184.
doi: 10.1093/jxb/eraa085 pmid: 32072171 |
| [39] |
Zheng JR, Yang XY, Ye JB, et al. Multiomics analysis provides new insights into the regulatory mechanism of carotenoid biosynthesis in yellow peach peel[J]. Mol Hortic, 2023, 3(1): 23.
doi: 10.1186/s43897-023-00070-3 pmid: 37919829 |
| [1] | 岳丽昕, 王清华, 刘泽洲, 孔素萍, 高莉敏. 基于转录组和WGCNA筛选大葱雄性不育相关基因[J]. 生物技术通报, 2024, 40(9): 212-224. |
| [2] | 聂祝欣, 郭瑾, 乔子洋, 李微薇, 张学燕, 刘春阳, 王静. 黑果枸杞不同发育时期果实花色苷合成的转录组分析[J]. 生物技术通报, 2024, 40(8): 106-117. |
| [3] | 周麟, 黄顺满, 苏文坤, 姚响, 屈燕. 滇山茶bHLH基因家族鉴定及花色形成相关基因筛选[J]. 生物技术通报, 2024, 40(8): 142-151. |
| [4] | 王睿, 戚继. 整合组织学图像信息增强空间转录组细胞聚类的分辨率[J]. 生物技术通报, 2024, 40(8): 39-46. |
| [5] | 高萌萌, 赵天宇, 焦馨悦, 林春晶, 关哲允, 丁孝羊, 孙妍妍, 张春宝. 大豆细胞质雄性不育系及其恢复系的比较转录组分析[J]. 生物技术通报, 2024, 40(7): 137-149. |
| [6] | 廖杨梅, 赵国春, 翁学煌, 贾黎明, 陈仲. 无患子雄性不育品种‘琦蕊’不同发育时期雄花转录组分析[J]. 生物技术通报, 2024, 40(7): 197-206. |
| [7] | 虞昕磊, 何结望, 林国平, 李金海, 王大爱, 袁跃斌, 刘圣高, 李志豪, 陶德欣. 夏冬两季发酵雪茄烟叶的代谢组差异分析[J]. 生物技术通报, 2024, 40(6): 260-270. |
| [8] | 秦健, 李振月, 何浪, 李俊玲, 张昊, 杜荣. 肌源性细胞分化的单细胞转录谱变化及细胞间通讯分析[J]. 生物技术通报, 2024, 40(6): 330-342. |
| [9] | 白志元, 徐菲, 杨午, 王明贵, 杨玉花, 张海平, 张瑞军. 大豆细胞质雄性不育弱恢复型杂种F1育性转变的转录组分析[J]. 生物技术通报, 2024, 40(6): 134-142. |
| [10] | 吴迪, 游小凤, 郑亦铮, 林楠, 张燕燕, 魏艺聪. 草珊瑚中类胡萝卜素合成的内源激素调控机制分析[J]. 生物技术通报, 2024, 40(5): 203-214. |
| [11] | 郭纯, 宋桂梅, 闫艳, 邸鹏, 王英平. 西洋参bZIP基因家族全基因组鉴定和表达分析[J]. 生物技术通报, 2024, 40(4): 167-178. |
| [12] | 钟匀, 林春, 刘正杰, 董陈文华, 毛自朝, 李兴玉. 芦笋皂苷合成相关糖基转移酶基因克隆及原核表达分析[J]. 生物技术通报, 2024, 40(4): 255-263. |
| [13] | 杨淇, 魏子迪, 宋娟, 童堃, 杨柳, 王佳涵, 刘海燕, 栾维江, 马轩. 水稻组蛋白H1三突变体的创建和转录组学分析[J]. 生物技术通报, 2024, 40(4): 85-96. |
| [14] | 谢倩, 江来, 贺进, 刘玲玲, 丁明月, 陈清西. 不同鲜食品质橄榄果实转录组测序及酚类代谢途径相关调控基因挖掘[J]. 生物技术通报, 2024, 40(3): 215-228. |
| [15] | 许沛冬, 易剑锋, 陈迪, 陈浩, 谢丙炎, 赵文军. 组学技术在生防芽胞杆菌的应用进展[J]. 生物技术通报, 2024, 40(10): 208-220. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||