








生物技术通报 ›› 2025, Vol. 41 ›› Issue (9): 265-276.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0364
• 研究报告 • 上一篇
吕镇1,2( ), 甘恬1, 霍思羽1, 赵晨笛1, 赵梦瑶1, 李亚涛1, 马玉超1(
), 甘恬1, 霍思羽1, 赵晨笛1, 赵梦瑶1, 李亚涛1, 马玉超1( ), 耿玉清2(
), 耿玉清2( )
)
收稿日期:2025-04-06
出版日期:2025-09-26
发布日期:2025-09-24
通讯作者:
马玉超,女,副教授,研究方向 :微生物代谢工程;E-mail: mayuchao@bifu.edu.cn作者简介:吕镇,男,硕士研究生,研究方向 :土壤微生物;E-mail: Lz1915@bjfu.edu.cn
基金资助:
LYU Zhen1,2( ), GAN Tian1, HUO Si-yu1, ZHAO Chen-di1, ZHAO Meng-yao1, LI Ya-tao1, MA Yu-chao1(
), GAN Tian1, HUO Si-yu1, ZHAO Chen-di1, ZHAO Meng-yao1, LI Ya-tao1, MA Yu-chao1( ), GENG Yu-qing2(
), GENG Yu-qing2( )
)
Received:2025-04-06
Published:2025-09-26
Online:2025-09-24
摘要:
目的 筛选surfactin高产菌株,从基因组水平了解菌株的遗传背景和surfactin的生物合成过程,探索surfactin的植物促生功能。 方法 利用选择性分离技术从植物根际土中筛选surfactin产生菌;分别采用高效液相色谱和MALDI-TOF质谱对surfactin进行定量和结构类型分析;利用Illumina 结合 PacBio三代测序平台进行全基因组测序,利用生物信息学进行分类鉴定、功能基因注释、次级代谢基因簇和surfactin的生物合成过程分析;利用盆栽实验检测surfactin对植物幼苗的促生功能。 结果 从采集自山西大同杨树根际土壤中筛选到排油能力和抑菌能力强的菌株C5A-1,所产surfactin结构类型为C14-和C15-surfactin A,产量为1 208.16 mg/L。C5A-1与Bacillus velezensis FZB42的基因组平均核苷酸一致性(ANI)和数字DNA-DNA杂交值分别为98.35%和85.4%。C5A-1的基因组大小为3 929 585 bp,GC含量为46.5%,共编码3 747个基因,包含12个次级代谢产物基因簇。surfactin生物合成基因簇包含了核心基因srfAA、srfAB和srfAC。C5A-1所产surfactin能够显著提高毛白杨、玉米和大豆的株高、茎粗、地上和地下部的干重。 结论 C5A-1为贝莱斯芽胞杆菌的一株新菌株,所产C14-和C15-surfactin A具有显著的植物促生功能,为代谢工程法改善surfactin产量提供了优良底盘细胞工厂,为surfactin在农林业的实际应用提供了理论基础。
吕镇, 甘恬, 霍思羽, 赵晨笛, 赵梦瑶, 李亚涛, 马玉超, 耿玉清. 产Surfactin贝莱斯芽胞杆菌C5A-1的鉴定和所产Surfactin对植物的促生效果[J]. 生物技术通报, 2025, 41(9): 265-276.
LYU Zhen, GAN Tian, HUO Si-yu, ZHAO Chen-di, ZHAO Meng-yao, LI Ya-tao, MA Yu-chao, GENG Yu-qing. Identification of Surfactin-producing Bacillus Velezensis C5A-1 and Evaluation of the Plant Growth-promoting Effects of Its Surfactin[J]. Biotechnology Bulletin, 2025, 41(9): 265-276.
样品名称 Sample name | 采样地 Sampling location | 植物种类 Plant species | 经纬度 Longitude and latitude |
|---|---|---|---|
| A5A | 湖南省岳阳市 | 番茄 | 112°90′E 28°68′N |
| A5C | 云南省丽江市 | 苹果 | 100°25′E 26°86′N |
| C5A | 山西省大同市 | 杨树 | 113°20′E 40°3′N |
| A6A | 北京市昌平区 | 黄豆 | 116°25′E 40°7′N |
| A6B | 湖北省仙桃市 | 白萝卜 | 112°20′E 30°18′N |
| A6D | 湖南省常德市 | 蒜苗 | 108°93’E 34°23’N |
表1 采样地情况
Table 1 Sampling sites
样品名称 Sample name | 采样地 Sampling location | 植物种类 Plant species | 经纬度 Longitude and latitude |
|---|---|---|---|
| A5A | 湖南省岳阳市 | 番茄 | 112°90′E 28°68′N |
| A5C | 云南省丽江市 | 苹果 | 100°25′E 26°86′N |
| C5A | 山西省大同市 | 杨树 | 113°20′E 40°3′N |
| A6A | 北京市昌平区 | 黄豆 | 116°25′E 40°7′N |
| A6B | 湖北省仙桃市 | 白萝卜 | 112°20′E 30°18′N |
| A6D | 湖南省常德市 | 蒜苗 | 108°93’E 34°23’N |

图1 Surfactin产生菌的筛选A:分离菌株的排油能力;B:分离菌株对立枯丝核菌的抑菌活性
Fig. 1 Screening of surfactin-producing strainsA: Oil-displacing e capacity of the isolated strains. B: Inhibitory activity of the isolated strains against R. solani
| Cluster | Type | Start | End | Similar cluster | Similarity (%) |
|---|---|---|---|---|---|
| 1 | NRPS | 323409 | 387387 | Surfactin | 82 |
| 2 | PKS-like | 923849 | 926504 | Butirosin A/B | 7 |
| 3 | Terpene | 1049972 | 1067381 | - | - |
| 4 | Lanthipeptide-class-ii | 1188370 | 1217259 | - | - |
| 5 | TransAT-PKS | 1383378 | 1471714 | Macrolactin H | 100 |
| 6 | TransAT-PKS | 1691242 | 1791808 | Bacillaene | 100 |
| 7 | NRPS | 1865549 | 1999860 | Fengycin | 100 |
| 8 | Terpene | 2028497 | 2025381 | - | - |
| 9 | T3PKS | 2113698 | 2154799 | - | - |
| 10 | TransAT-PKS | 2282174 | 2375967 | Difficidin | 100 |
| 11 | NRPS | 300670 | 3052462 | Bacillibactin | 100 |
| 12 | Other | 3588771 | 3630190 | Bacilysin | 100 |
表2 C5A-1全基因组次级代谢基因簇分析
Table 2 Genome-wide secondary metabolite gene clusters from C5A-1
| Cluster | Type | Start | End | Similar cluster | Similarity (%) |
|---|---|---|---|---|---|
| 1 | NRPS | 323409 | 387387 | Surfactin | 82 |
| 2 | PKS-like | 923849 | 926504 | Butirosin A/B | 7 |
| 3 | Terpene | 1049972 | 1067381 | - | - |
| 4 | Lanthipeptide-class-ii | 1188370 | 1217259 | - | - |
| 5 | TransAT-PKS | 1383378 | 1471714 | Macrolactin H | 100 |
| 6 | TransAT-PKS | 1691242 | 1791808 | Bacillaene | 100 |
| 7 | NRPS | 1865549 | 1999860 | Fengycin | 100 |
| 8 | Terpene | 2028497 | 2025381 | - | - |
| 9 | T3PKS | 2113698 | 2154799 | - | - |
| 10 | TransAT-PKS | 2282174 | 2375967 | Difficidin | 100 |
| 11 | NRPS | 300670 | 3052462 | Bacillibactin | 100 |
| 12 | Other | 3588771 | 3630190 | Bacilysin | 100 |
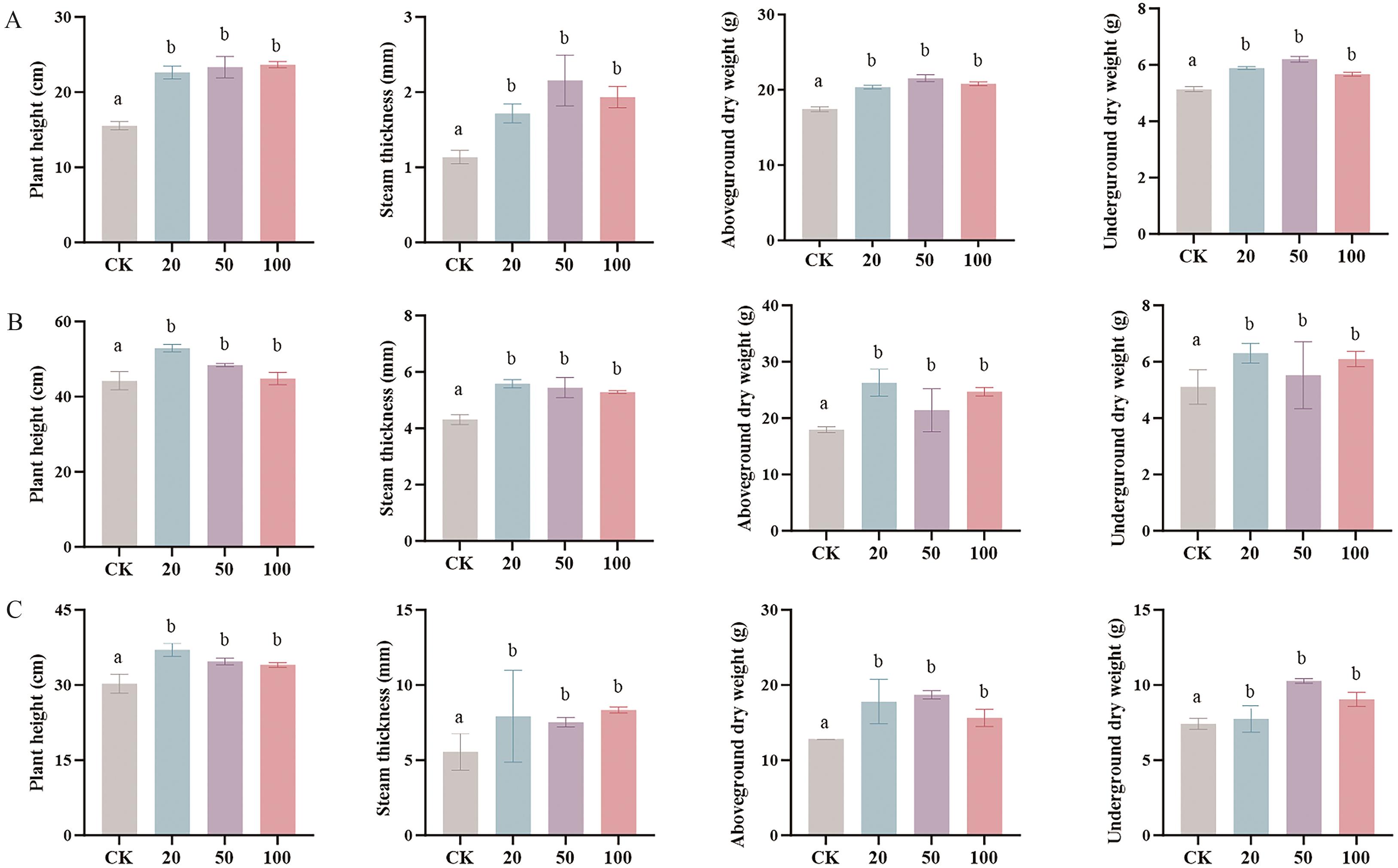
图8 C5A-1所产的不同浓度surfactin对植物的促生功能A:毛白杨 84K:B:大豆;C:玉米。不同小写字母代表处理间差异显著(P<0.05)
Fig. 8 Plant-growth promoting by different concentrations of surfactin from C5A-1A: Populus tomentosa 84K. B: Glycine max. C: Zea mays. Different lowercase letters indicate significant differences between treatments (P<0.05)
| [1] | Sreedharan SM, Rishi N, Singh R. Microbial lipopeptides: Properties, mechanics and engineering for novel lipopeptides [J]. Microbiol Res, 2023, 271: 127363. |
| [2] | 李道明, 王瑛, 陈超, 等. 芽胞杆菌几种重要抗菌脂肽研究进展 [J]. 生物工程学报, 2022, 38(5): 1768-1783. |
| Li DM, Wang Y, Chen C, et al. Advances in several important antimicrobial lipopeptids from Bacillus spp [J]. Chin J Biotechnol, 2022, 38(5): 1768-1783. | |
| [3] | Hu FX, Liu YY, Li S. Rational strain improvement for surfactin production: enhancing the yield and generating novel structures [J]. Microb Cell Fact, 2019, 18(1): 42. |
| [4] | Augustyn AR, Pott RWM, Tadie M. The interactions of the biosurfactant surfactin in coal flotation [J]. Colloids Surf A Physicochem Eng Aspects, 2021, 627: 127122. |
| [5] | Théatre A, Cano-Prieto C, Bartolini M, et al. The surfactin-like lipopeptides from Bacillus spp. natural biodiversity and synthetic biology for a broader application range [J]. Front Bioeng Biotechnol, 2021, 9: 623701. |
| [6] | Saiyam D, Dubey A, Malla MA, et al. Lipopeptides from Bacillus: unveiling biotechnological prospects-sources, properties, and diverse applications [J]. Braz J Microbiol, 2024, 55(1): 281-295. |
| [7] | Munusamy S, Conde R, Bertrand B, et al. Biophysical approaches for exploring lipopeptide-lipid interactions [J]. Biochimie, 2020, 170: 173-202. |
| [8] | Chen XY, Lu YJ, Shan MY, et al. A mini-review: mechanism of antimicrobial action and application of surfactin [J]. World J Microbiol Biotechnol, 2022, 38(8): 143. |
| [9] | Laird M, Piccoli D, Weselowski B, et al. Surfactin-producing Bacillus velezensis 1B-23 and Bacillus sp. 1D-12 protect tomato against bacterial canker caused by Clavibacter michiganensis subsp. michiganensis [J]. J Plant Pathol, 2020, 102(2): 451-458. |
| [10] | Ali SAM, Sayyed RZ, Mir MI, et al. Induction of systemic resistance in maize and antibiofilm activity of surfactin from Bacillus velezensis MS20 [J]. Front Microbiol, 2022, 13: 879739. |
| [11] | Wang YY, Zhang CY, Liang J, et al. Surfactin and fengycin B extracted from Bacillus pumilus W-7 provide protection against potato late blight via distinct and synergistic mechanisms [J]. Appl Microbiol Biotechnol, 2020, 104(17): 7467-7481. |
| [12] | Wang MM, Yu HM, Li X, et al. Single-gene regulated non-spore-forming Bacillus subtilis: Construction, transcriptome responses, and applications for producing enzymes and surfactin [J]. Metab Eng, 2020, 62: 235-248. |
| [13] | 王俊芳, 黄秋斌, 张飘丹, 等. Surfactin的结构、生物合成及其在生物防治中的作用 [J]. 生物技术通报, 2024, 40(1): 100-112. |
| Wang JF, Huang QB, Zhang PD, et al. The structure, biosynthesis of surfactin and its role in biological control [J]. Biotechnol Bull, 2024, 40(1): 100-112. | |
| [14] | Shu ZZ, Yan PF, Huang LR, et al. Improvement of interfacial, antioxidant, and emulsifying properties of pectin by grafting surfactin [J]. Int J Biol Macromol, 2025, 307: 142091. |
| [15] | Richter M, Rosselló-Móra R, Oliver Glöckner F, et al. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison [J]. Bioinformatics, 2016, 32(6): 929-931. |
| [16] | Luo RB, Liu BH, Xie YL, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler [J]. Gigascience, 2012, 1(1): 18. |
| [17] | Krzywinski M, Schein J, Birol I, et al. Circos: an information aesthetic for comparative genomics [J]. Genome Res, 2009, 19(9): 1639-1645. |
| [18] | Lombard V, Golaconda Ramulu H, Drula E, et al. The carbohydrate-active enzymes database (CAZy) in 2013 [J]. Nucleic Acids Res, 2014, 42(Database issue): D490-D495. |
| [19] | Blin K, Shaw S, Augustijn HE, et al. antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation [J]. Nucleic Acids Res, 2023, 51(W1): W46-W50. |
| [20] | Balleux G, Höfte M, Arguelles-Arias A, et al. Bacillus lipopeptides as key players in rhizosphere chemical ecology [J]. Trends Microbiol, 2025, 33(1): 80-95. |
| [21] | 李光月, 胡文锋, 李雪玲. 复合诱变改善枯草芽胞杆菌表面活性素的抗菌能力 [J]. 农业生物技术学报, 2023, 31(7): 1488-1500. |
| Li GY, Hu WF, Li XL. Enhancing the antimicrobial capacity of surfactin in Bacillus subtilis through composite mutagenesis [J]. Journal of Agricultural Biotechnology, 2023, 31(7): 1488-1500. | |
| [22] | Stoll A, Salvatierra-Martínez R, González M, et al. The role of surfactin production by Bacillus velezensis on colonization, biofilm formation on tomato root and leaf surfaces and subsequent protection (ISR) against Botrytis cinerea [J]. Microorganisms, 2021, 9(11): 2251. |
| [23] | Bochynek M, Lewińska A, Witwicki M, et al. Formation and structural features of micelles formed by surfactin homologues [J]. Front Bioeng Biotechnol, 2023, 11: 1211319. |
| [24] | Klapper M, Braga D, Lackner G, et al. Bacterial alkaloid biosynthesis: structural diversity via a minimalistic nonribosomal peptide synthetase [J]. Cell Chem Biol, 2018, 25(6): 659-665.e9. |
| [25] | Fortinez CM, Bloudoff K, Harrigan C, et al. Structures and function of a tailoring oxidase in complex with a nonribosomal peptide synthetase module [J]. Nat Commun, 2022, 13(1): 548. |
| [26] | Xu Y, Wu JY, Liu QJ, et al. Genome-wide identification and evolutionary analyses of SrfA operon genes in Bacillus [J]. Genes, 2023, 14(2): 422. |
| [27] | Yaseen Y, Diop A, Gancel F, et al. Polynucleotide phosphorylase is involved in the control of lipopeptide fengycin production in Bacillus subtilis [J]. Arch Microbiol, 2018, 200(5): 783-791. |
| [28] | Zhen C, Ge XF, Lu YT, et al. Chemical structure, properties and potential applications of surfactin, as well as advanced strategies for improving its microbial production [J]. AIMS Microbiol, 2023, 9(2): 195-217. |
| [29] | Sharma N, Singhvi R. Effects of chemical fertilizers and pesticides on human health and environment: a review [J]. Intern Jour Agricul, Environ And Biotech, 2017, 10(6): 675. |
| [30] | Kumar A, Singh SK, Kant C, et al. Microbial biosurfactant: a new frontier for sustainable agriculture and pharmaceutical industries [J]. Antioxidants, 2021, 10(9): 1472. |
| [31] | Karamchandani BM, Pawar AA, Pawar SS, et al. Biosurfactants’ multifarious functional potential for sustainable agricultural practices [J]. Front Bioeng Biotechnol, 2022, 10: 1047279. |
| [32] | 杨柳, 邓杰勇, 王青青, 等. 表面活性素对不结球白菜叶片生长和硼吸收的促进 [J]. 江苏农业学报, 2016, 32(5): 1134-1140. |
| Yang L, Deng JY, Wang QQ, et al. Improved uptake of boron and growth in Chinese cabbage leaves by surfactin [J]. Jiangsu J Agric Sci, 2016, 32(5): 1134-1140. |
| [1] | 石艳华, 李朔, 高玉珠, 郑保坤, 朱杰华, 张岱, 杨志辉. 贝莱斯芽胞杆菌NZ-4挥发性有机物的促生作用及其活性成分分析[J]. 生物技术通报, 2025, 41(8): 300-310. |
| [2] | 张津浩, 邓辉, 张清壮, 陶禹, 周池, 李鑫. 贝莱斯芽胞杆菌XY40-1对百合球茎生长、品质及镉含量的调控作用[J]. 生物技术通报, 2025, 41(7): 281-291. |
| [3] | 张慧, 卢文才, 王冬, 刘倩, 马连杰. 一株高产吲哚乙酸的Bacillus cereus YT2-1C的鉴定及促生作用[J]. 生物技术通报, 2025, 41(5): 300-309. |
| [4] | 吴泽银, 黄晨阳, 赵梦然, 张利姣, 姚方杰. 短柄白黄侧耳CCMSSC 04611基因组特异性分析[J]. 生物技术通报, 2025, 41(5): 320-332. |
| [5] | 张婷, 万雨欣, 徐伟慧, 王志刚, 陈文晶, 胡云龙. 一株玉米根际促生菌Leclercia adecarboxylata LN01促生效果研究及其基因组分析[J]. 生物技术通报, 2025, 41(1): 263-275. |
| [6] | 刘倩, 马连杰, 张慧, 王冬, 范茂, 廖敦秀, 赵正武, 卢文才. 辣椒炭疽病生防菌株TN2的筛选鉴定与抑菌效果[J]. 生物技术通报, 2025, 41(1): 287-297. |
| [7] | 周江鸿, 夏菲, 仲丽, 仇兰芬, 李广, 刘倩, 张国锋, 邵金丽, 李娜, 车少臣. 黄栌枯萎病拮抗细菌CCBC3-3-1的全基因组测序及比较基因组分析[J]. 生物技术通报, 2024, 40(7): 235-246. |
| [8] | 田彤彤, 葛家振, 高鹏程, 李学瑞, 宋国栋, 郑福英, 储岳峰. 绵羊肺炎支原体GH3-3株全基因组测序及生物信息学分析[J]. 生物技术通报, 2024, 40(7): 323-334. |
| [9] | 孙亚楠, 王春雪, 王欣, 杜秉海, 刘凯, 汪城墙. 萎缩芽孢杆菌CNY01的生防特性及其对玉米的抗盐促生作用[J]. 生物技术通报, 2024, 40(5): 248-260. |
| [10] | 许沛冬, 易剑锋, 陈迪, 潘磊, 谢丙炎, 赵文军. 贝莱斯芽孢杆菌生防次级代谢产物研究进展[J]. 生物技术通报, 2024, 40(3): 75-88. |
| [11] | 王梓, 石金川, 王永强, 孙淼, 孟令浩, 耿超, 刘锴. 牛源荚膜A型、D型多杀性巴氏杆菌的全基因组测序及基因组进化分析[J]. 生物技术通报, 2024, 40(12): 282-290. |
| [12] | 王俊芳, 黄秋斌, 张飘丹, 张彭湃. Surfactin的结构、生物合成及其在生物防治中的作用[J]. 生物技术通报, 2024, 40(1): 100-112. |
| [13] | 王腾辉, 葛雯冬, 罗雅方, 范震宇, 王玉书. 基于极端混合池(BSA)全基因组重测序的羽衣甘蓝白色叶基因定位[J]. 生物技术通报, 2023, 39(9): 176-182. |
| [14] | 方澜, 黎妍妍, 江健伟, 成胜, 孙正祥, 周燚. 盘龙参内生真菌胞内细菌7-2H的分离鉴定和促生特性研究[J]. 生物技术通报, 2023, 39(8): 272-282. |
| [15] | 郭少华, 毛会丽, 刘征权, 付美媛, 赵平原, 马文博, 李旭东, 关建义. 一株鱼源致病性嗜水气单胞菌XDMG的全基因组测序及比较基因组分析[J]. 生物技术通报, 2023, 39(8): 291-306. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||