








生物技术通报 ›› 2025, Vol. 41 ›› Issue (11): 212-220.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0527
• 技术与方法 • 上一篇
桑世博( ), 李俐, 张枫源, 孙朗, 谌能双, 程聪, 任燕萍, 马丽, 张桦(
), 李俐, 张枫源, 孙朗, 谌能双, 程聪, 任燕萍, 马丽, 张桦( )
)
收稿日期:2025-05-22
出版日期:2025-11-26
发布日期:2025-12-09
通讯作者:
张桦,女,博士,教授,研究方向 :植物生物技术、荒漠植物生态适应性分子机制;E-mail: hazelzhang@163.com作者简介:桑世博,男,硕士研究生,研究方向 :荒漠植物抗逆机制;E-mail: 927810653@qq.com
基金资助:
SANG Shi-bo( ), LI Li, ZHANG Feng-yuan, SUN Lang, SHEN Neng-shuang, CHENG Cong, REN Yan-ping, MA Li, ZHANG Hua(
), LI Li, ZHANG Feng-yuan, SUN Lang, SHEN Neng-shuang, CHENG Cong, REN Yan-ping, MA Li, ZHANG Hua( )
)
Received:2025-05-22
Published:2025-11-26
Online:2025-12-09
摘要:
目的 建立一套适用于梭梭的病毒诱导基因沉默(virus-mediated gene silencing, VIGS)体系,并验证其有效性。 方法 以梭梭幼苗为实验材料,选取CLA1(Cloroplastosalterados1)基因为指示基因,构建重组病毒载体,并将其转化至GV3101农杆菌中。采用根吸收法侵染四周龄梭梭根部,探索不同侵染液对烟草脆裂病毒(TRV)诱导梭梭基因沉默的影响。利用该体系验证梭梭HaNAC3基因是否在盐胁迫中发挥正调控作用。 结果 采用农杆菌介导的根吸收法和烟草匀浆介导的根吸收法均成功诱导梭梭幼苗产生白化表型。相较于空载对照组,沉默效率分别为57.93%和77.66%,HaCLA1基因相对表达量均显著降低,其中烟草匀浆侵染组的沉默效果更为显著,从而初步建立了梭梭VIGS体系。我们利用该体系将HaCLA1基因作为阳性对照,成功沉默了梭梭HaNAC3耐盐基因。与对照组相比,基因沉默后的梭梭植株耐盐性显著减弱。 结论 建立了梭梭基因沉默体系,利用烟草复制匀浆二次侵染可以提高基因沉默效率,同时利用该体系验证了梭梭HaNAC3耐盐基因功能。
桑世博, 李俐, 张枫源, 孙朗, 谌能双, 程聪, 任燕萍, 马丽, 张桦. TRV介导的梭梭基因沉默体系构建与验证[J]. 生物技术通报, 2025, 41(11): 212-220.
SANG Shi-bo, LI Li, ZHANG Feng-yuan, SUN Lang, SHEN Neng-shuang, CHENG Cong, REN Yan-ping, MA Li, ZHANG Hua. Construction and Validation of the TRV-mediated Gene Silencing System in Haloxylon ammodendron[J]. Biotechnology Bulletin, 2025, 41(11): 212-220.
引物名称 Name of primer | 引物序列 Sequence of primer (5′-3′) | 用途 Application |
|---|---|---|
| pTRV2-HaCLA1-F | GTGAGTAAGGTTACCGAATTCAGGAAGACCTCAGGGCTAGCA | 沉默载体构建 Silent vector construction |
| pTRV2-HaCLA1-R | GAGACGCGTGAGCTCGGTACCCGTCCACTGGACCAATGTAGTACA | |
| pTRV2-HaNAC3-F | TGAGTAAGGTTACCGAATTCGTACTACGAGGGAAAATATCCATCATC | |
| pTRV2-HaNAC3-R | GAGACGCGTGAGCTCGGTACCTCATCTTGATCATCATTGTCTTCCTC | |
| 18SrRNA-F | CTCTGCCGTTGCTCTGATGAT | 基因相对表达量分析 Analysis of relative gene expressions |
| 18SrRNA-R | CCTTGGATGTGGTAGCCGTTC | |
| RT-qPCR-HaNAC3-F | ATCAAAACAGCCAGATGTCA | |
| RT-qPCR-HaNAC3-R | GAAGAAAGCTCCACTTCGTTG | |
| RT-qPCR-HACLA1-F | CTTTAGGGAAGCTCCAAGCATT | |
| RT-qPCR-HACLA1-R | CGCAACCTTGTGTGTTTGTCC |
表1 试验中所用到的引物
Table 1 Primers used in the study
引物名称 Name of primer | 引物序列 Sequence of primer (5′-3′) | 用途 Application |
|---|---|---|
| pTRV2-HaCLA1-F | GTGAGTAAGGTTACCGAATTCAGGAAGACCTCAGGGCTAGCA | 沉默载体构建 Silent vector construction |
| pTRV2-HaCLA1-R | GAGACGCGTGAGCTCGGTACCCGTCCACTGGACCAATGTAGTACA | |
| pTRV2-HaNAC3-F | TGAGTAAGGTTACCGAATTCGTACTACGAGGGAAAATATCCATCATC | |
| pTRV2-HaNAC3-R | GAGACGCGTGAGCTCGGTACCTCATCTTGATCATCATTGTCTTCCTC | |
| 18SrRNA-F | CTCTGCCGTTGCTCTGATGAT | 基因相对表达量分析 Analysis of relative gene expressions |
| 18SrRNA-R | CCTTGGATGTGGTAGCCGTTC | |
| RT-qPCR-HaNAC3-F | ATCAAAACAGCCAGATGTCA | |
| RT-qPCR-HaNAC3-R | GAAGAAAGCTCCACTTCGTTG | |
| RT-qPCR-HACLA1-F | CTTTAGGGAAGCTCCAAGCATT | |
| RT-qPCR-HACLA1-R | CGCAACCTTGTGTGTTTGTCC |
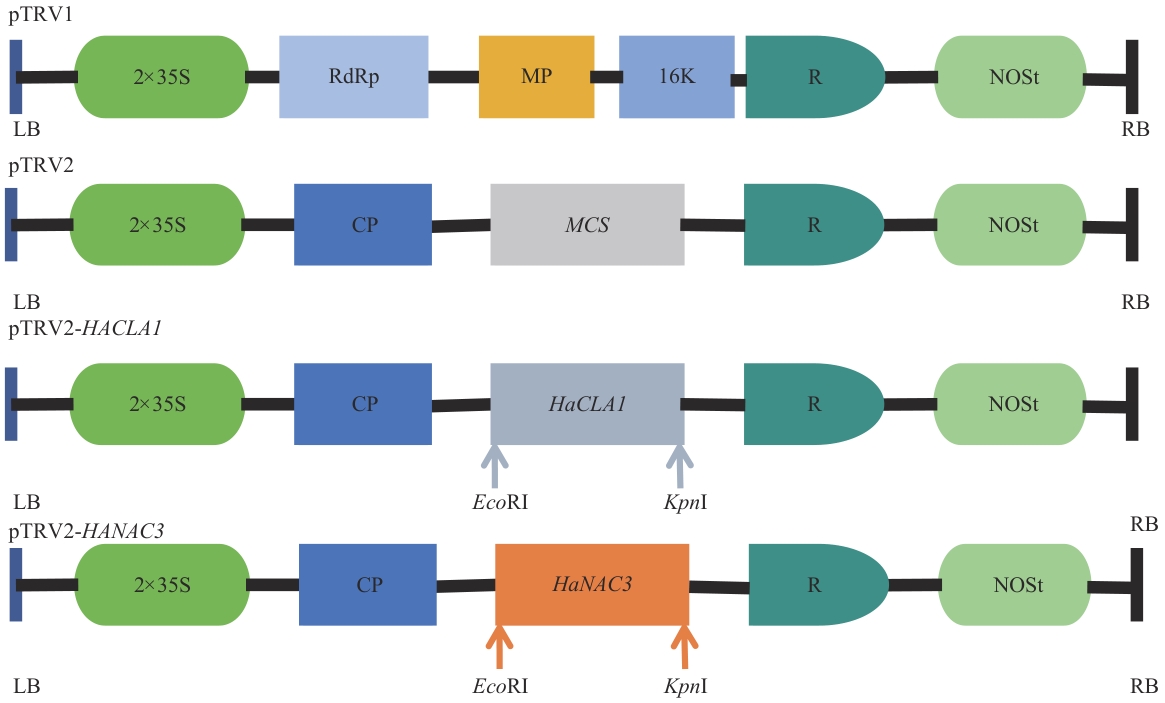
图1 pTRV1、 pTRV2、 pTRV2-HaCLA1、 pTRV2-HaNAC3载体示意图LB:左缘;RB:右缘;35S:CaMV 35S 启动子;R:自裂核酶;NOSt:NOS终止子;CP:病毒外壳蛋白
Fig. 1 Schematic diagram of pTRV1, pTRV2, pTRV2-HaCLA1and pTRV2-HaNAC3 vectorsLB: Left border. RB: Right border. 35S: CaMV 35S promoter. R: Self-cleaving ribozyme. NOSt: NOS terminator. CP: Viral capsid protein
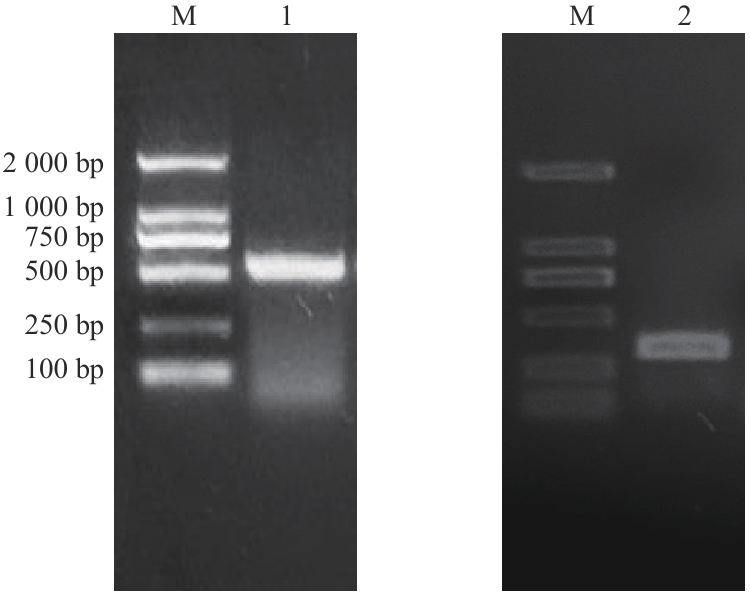
图3 基因沉默片段克隆琼脂糖凝胶电泳验证M:2 000 DNA marker;1:HaCLA1基因沉默片段;2:HaNAC3基因沉默片段
Fig. 3 Agarose gel electrophoresis verification of gene silencing fragment cloneM: 2 000 DNA marker. 1: HaCLA1 gene silencing fragment; 2: HaNAC3 gene silencing fragment

图4 重组病毒载体双酶切验证M:10 000 DNA Marker;1:pTRV2-HaCLA1载体双酶切;2:pTRV2-HaCLA1原始质粒电泳
Fig. 4 Recombinant viral vector double enzyme digestion and verificationM:10 000 DNA marker;1:double digestion of pTRV2-HaCLA1 vector;2:pTRV2-HaCLA1 original plasmid electrophoresis
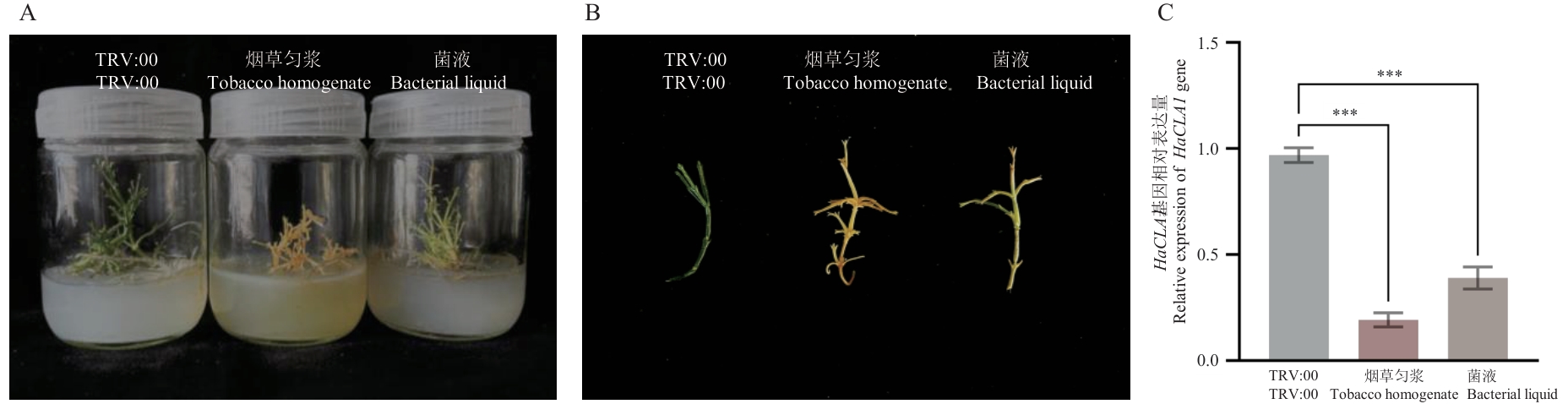
图5 不同侵染液下梭梭HaCLA1基因沉默表型及RT-qPCR分析结果(侵染12 d)A:HaCLA1基因沉默后梭梭整株沉默表型;B:HaCLA1基因沉默后梭梭同化枝沉默表型;C:HaCLA1基因的RT-qPCR分析结果(侵染12 d)
Fig. 5 Phenotype of HaCLA1 gene silencing and RT-qPCR analysis under different infection solutions (12 d of infestation)A:Whole-plant silencing phenotype of H. ammodendron after HaCLA1 gene silencing. B:Assimilating branch silencing phenotype of H. ammodendron after HaCLA1 gene silencing. C: Results of RT-qPCR analysis of HaCLA1 gene (12 d of infestation). ***P<0.001
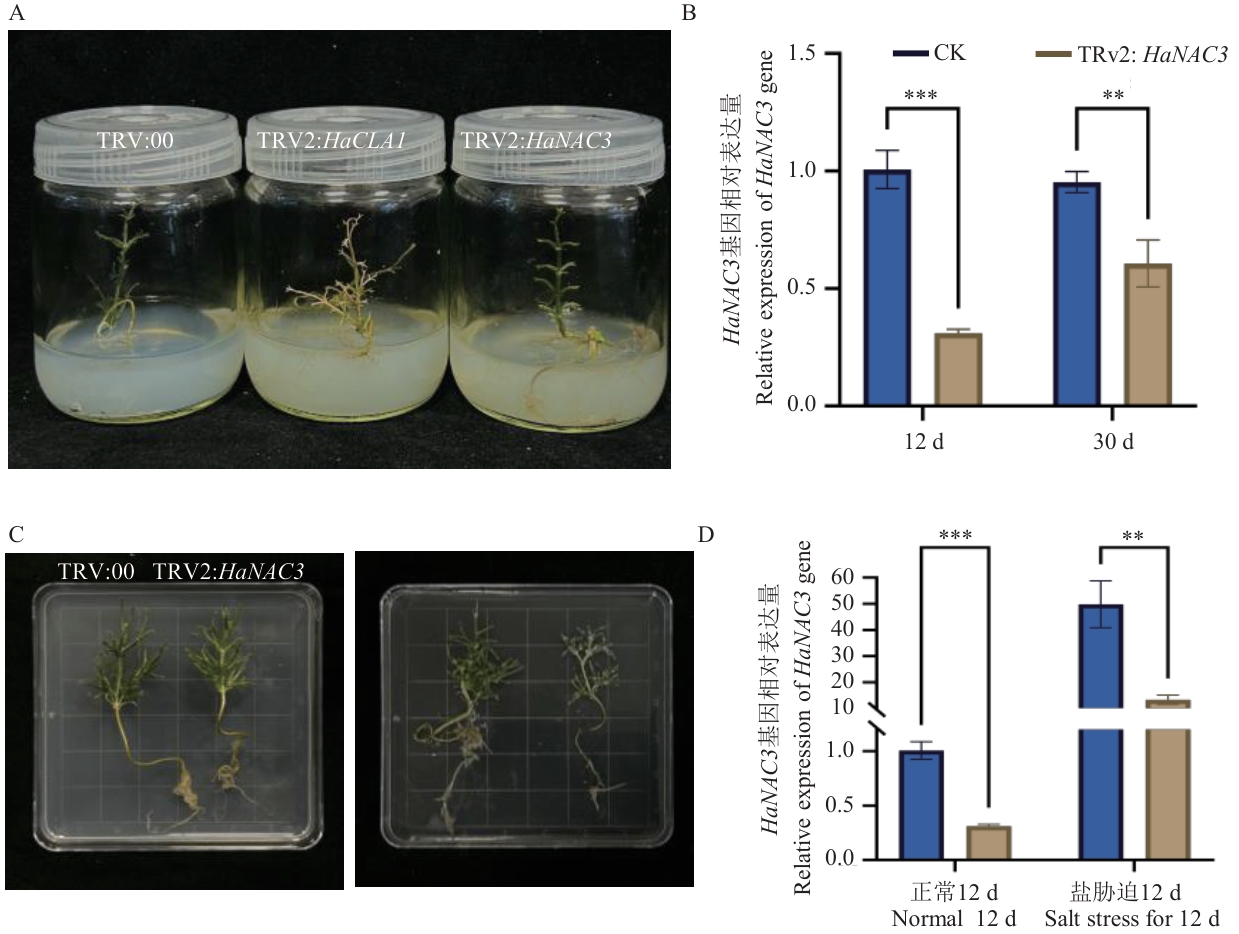
图6 梭梭VIGS体系应用A:梭梭HaNAC3基因沉默植株表型(TRV:00:空载体阴性对照组;TRV2:HaCLA1;阳性对照组;TRV2:HaNAC3:实验组);B:梭梭12 d、30 d后HaNAC3基因相对表达量;C:梭梭正常情况下和盐胁迫12 d后植株表型;D:梭梭正常情况和盐胁迫12 d后HaNAC3基因相对表达量,** P<0.01水平上存在显著差异;*** P<0.001水平上存在显著差异
Fig. 6 Application of the H. ammodendron VIGS systemA: Phenotypic changes of HaNAC3 gene silencing in H. ammodendron. (TRV:00:Empty vector negative control group;TRV2:HaCLA1: Positive control group;TRV2:HaNAC3: Experimental group). B: Relative expression of the HaNAC3 gene in H. ammodendron after 12 d and 30 d. C: Phenotypic characteristics of H. ammodendron plants under normal conditions and after 12 d of salt stress. D: Relative expressions of the HaNAC3 gene in H. ammodendron under normal conditions and after 12 d of salt stress. ** indicates significant differences at the P<0.01, and *** indicates significant differences at the P<0.001
| [1] | Song J, Feng G, Tian CY, et al. Osmotic adjustment traits of Suaeda physophora, Haloxylon ammodendron and Haloxylon persicum in field or controlled conditions [J]. Plant Sci, 2006, 170(1): 113-119. |
| [2] | 张丹, 马松梅, 魏博, 等. 中国梭梭属植物历史分布格局及其驱动机制 [J]. 生物多样性, 2022, 30(1):42-51 |
| Zhang D, Ma SM, Wei B, et al. Historical distribution pattern and driving mechanism of Haloxylon in China [J]. Biodivers Sci, 2022, 30(1):42-51 | |
| [3] | 郑旭, 张倪斌, 孙桂丽, 等. 吐鲁番市高昌区不同固沙植物的防风固沙效益 [J]. 东北林业大学学报, 2024, 52(7): 44-50. |
| Zheng X, Zhang NB, Sun GL, et al. Wind prevention and sand-fixation benefits of different sand-fixing plants in Gaochang district, Turpan City [J]. J Northeast For Univ, 2024, 52(7): 44-50. | |
| [4] | 沈亮, 徐荣, 刘赛, 等. 肉苁蓉寄主梭梭根际土壤微生物种类及群落结构特征 [J]. 生态学报, 2016, 36(13): 3933-3942. |
| Shen L, Xu R, Liu S, et al. Characteristics of microbial community structure in rhizosphere soil of Haloxylon ammodendron [J]. Acta Ecol Sin, 2016, 36(13): 3933-3942. | |
| [5] | 许丽, 刘永刚, 王菊莲, 等. 石羊河下游荒漠区雨养梭梭柠条人工林健康评价 [J]. 防护林科技, 2025(1): 70-77. |
| Xu L, Liu YG, Wang JL, et al. Health evaluation of rain-fed Haloxylon ammodendron and Caragana korshinskii plantation in desert area of lower reaches of Shiyang River [J]. Prot For Sci Technol, 2025(1): 70-77. | |
| [6] | Liang JS, Liu XS, Xu L, et al. A novel NAC transcription factor from Haloxylon ammodendron promotes reproductive growth in Arabidopsis thaliana under drought stress [J]. Environ Exp Bot, 2024, 228: 106043. |
| [7] | Senthil-Kumar M, Mysore KS. New dimensions for VIGS in plant functional genomics [J]. Trends Plant Sci, 2011, 16(12): 656-665. |
| [8] | 潘多, 张嵩玥, 刘芳伊, 等. 病毒诱导的基因沉默技术用于植物色素代谢机制的研究进展 [J]. 生物工程学报, 2023, 39(7): 2579-2599. |
| Pan D, Zhang SY, Liu FY, et al. Application of virus-induced gene silencing technology to investigate the phytochrome metabolism mechanism: a review [J]. Chin J Biotechnol, 2023, 39(7): 2579-2599. | |
| [9] | Rosa C, Kuo YW, Wuriyanghan H, et al. RNA interference mechanisms and applications in plant pathology [J]. Annu Rev Phytopathol, 2018, 56: 581-610. |
| [10] | Yang W, Chen XY, Chen JH, et al. Virus-induced gene silencing in the tea plant (Camellia sinensis) [J]. Plants, 2023, 12(17): 3162. |
| [11] | 李瑞雪, 王钰婷, 胡飞, 等. 桑树PDS基因VIGS转化体系的构建与鉴定 [J]. 南方农业学报, 2018, 49(7): 1432-1438. |
| Li RX, Wang YT, Hu F, et al. VIGS transformation system construction and identification of gene PDS in mulberry [J]. J South Agric, 2018, 49(7): 1432-1438. | |
| [12] | Sasaki S, Yamagishi N, Yoshikawa N. Efficient virus-induced gene silencing in apple, pear and Japanese pear using Apple latent spherical virus vectors [J]. Plant Methods, 2011, 7(1): 15. |
| [13] | Cheuk A, Houde M. A rapid and efficient method for uniform gene expression using the barley stripe mosaic virus [J]. Plant Methods, 2017, 13: 24. |
| [14] | Mandel MA, Feldmann KA, Herrera-Estrella L, et al. CLA1 a novel gene required for chloroplast development, is highly conserved in evolution [J]. Plant J, 1996, 9(5): 649-658. |
| [15] | Senthil-Kumar M, Mysore KS. Tobacco rattle virus-based virus-induced gene silencing in Nicotiana benthamiana [J]. Nat Protoc, 2014, 9(7): 1549-1562. |
| [16] | Wang P, Man LJ, Ma L, et al. In vitro regeneration of Haloxylon ammodendron [J]. Not Sci Biol, 2023, 15(2): 11585. |
| [17] | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method [J]. Methods, 2001, 25(4): 402-408. |
| [18] | Chen PJ, Pang G, Cai F, et al. Strain improvement and genetic engineering of Trichoderma for industrial applications [M]//Encyclopedia of Mycology. Amsterdam: Elsevier, 2021: 505-517. |
| [19] | Azeez A, Bates PD. Self-incompatibility based functional genomics for rapid phenotypic characterization of seed metabolism genes [J]. Plant Biotechnol J, 2024, 22(10): 2688-2690. |
| [20] | Zhu YY, Yu XP, Wu J. CRISPR/Cas: a toolkit for plant disease diagnostics [J]. Trends Plant Sci, 2025, 30(3): 245-248. |
| [21] | Liu XS, Zong XF, Wu X, et al. Ectopic expression of NAC transcription factor HaNAC3 from Haloxylon ammodendron increased abiotic stress resistance in tobacco [J]. Planta, 2022, 256(6): 105. |
| [22] | Wang MC, Zhang L, Tong SF, et al. Chromosome-level genome assembly of a xerophytic plant, Haloxylon ammodendron [J]. DNA Res, 2022, 29(2): dsac006. |
| [23] | 张桦, 娜菲莎·木则帕尔, 任燕萍, 等.农杆菌介导的梭梭种子遗传转化体系构建的方法: CN118620955A [P]. 2024-09-10. |
| Zhang H, Nafeisha M, Ren YP, et al. Method for constructing a genetic transformation system of Haloxylon ammodendron seeds mediated by Agrobacterium: CN118620955A [P]. 2024-09-10. | |
| [24] | Cao YH, Ren W, Gao HJ, et al. HaASR2 from Haloxylon ammodendron confers drought and salt tolerance in plants [J]. Plant Sci, 2023, 328: 111572. |
| [25] | Navarro JA, Sanchez-Navarro JA, PALLAS V. Chapter One - Key checkpoints in the movement of plant viruses through the host [M] Advances in Virus Research: Vol. 104. Academic Press, 2019: 1-64. |
| [26] | Shi GY, Hao MY, Tian BM, et al. A newly established virus-induced gene silencing method via seed imbibition for functional genomics at early germination stages in cotton [J]. Ind Crops Prod, 2021, 172: 114040. |
| [27] | Scofield SR, Huang L, Brandt AS, et al. Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway [J]. Plant Physiol, 2005, 138(4): 2165-2173. |
| [28] | Kant R, Dasgupta I. Gene silencing approaches through virus-based vectors: speeding up functional genomics in monocots [J]. Plant Mol Biol, 2019, 100(1/2): 3-18. |
| [29] | Lu Y, Zeng FJ, Zhang ZH, et al. Differences in growth, ionomic and antioxidative enzymes system responded to neutral and alkali salt exposure in halophyte Haloxylon ammodendron seedlings [J]. Plant Physiol Biochem, 2025, 220: 109492. |
| [30] | Lü XP, Lü ZL, Zhang YM, et al. Lignin synthesis plays an essential role in the adaptation of Haloxylon ammodendron to adverse environments [J]. Int J Biol Macromol, 2025, 308: 142321. |
| [31] | 杨芳. 梭梭和白梭梭响应干旱的生理代谢和分子机制[D]. 乌鲁木齐: 新疆大学, 2022. |
| Yang F. Physiological metabolism and molecular mechanisms of Haloxylon ammodendron and Haloxylonpersicum in response to drought [D]. Urumqi: Xinjiang University, 2022 |
| [1] | 牛景萍, 赵婧, 郭茜, 王书宏, 赵晋忠, 杜维俊, 殷丛丛, 岳爱琴. 基于WGCNA鉴定大豆抗大豆花叶病毒NAC转录因子及其诱导表达分析[J]. 生物技术通报, 2025, 41(7): 95-105. |
| [2] | 张勇, 宋盛龙, 李永泰, 张新宇, 李艳军. 陆地棉GhSWEET9基因的克隆及抗黄萎病功能分析[J]. 生物技术通报, 2025, 41(6): 144-154. |
| [3] | 华炫, 田博雯, 周欣彤, 江梓涵, 王诗琦, 黄倩慧, 张健, 陈艳红. 旱柳SmERF B3-45的克隆及耐盐功能研究[J]. 生物技术通报, 2024, 40(12): 124-135. |
| [4] | 张怡, 张心如, 张金珂, 胡利宗, 上官欣欣, 郑晓红, 胡娟娟, 张聪聪, 穆桂清, 李成伟. 小麦镉胁迫响应基因TaMYB1的功能分析[J]. 生物技术通报, 2024, 40(1): 194-206. |
| [5] | 李文辰, 刘鑫, 康越, 李伟, 齐泽铮, 于璐, 王芳. TRV病毒诱导大豆基因沉默体系优化及应用[J]. 生物技术通报, 2023, 39(7): 143-150. |
| [6] | 冯策婷, 江律, 刘鑫颖, 罗乐, 潘会堂, 张启翔, 于超. 单叶蔷薇NAC基因家族鉴定及干旱胁迫响应分析[J]. 生物技术通报, 2023, 39(11): 283-296. |
| [7] | 张玉娟, 黎冬华, 宫慧慧, 崔新晓, 高春华, 张秀荣, 游均, 赵军胜. 芝麻NAC转录因子基因SiNAC77的克隆及耐盐功能分析[J]. 生物技术通报, 2023, 39(11): 308-317. |
| [8] | 李秀青, 胡子曜, 雷建峰, 代培红, 刘超, 邓嘉辉, 刘敏, 孙玲, 刘晓东, 李月. 棉花黄萎病抗性相关基因GhTIFY9的克隆与功能分析[J]. 生物技术通报, 2022, 38(8): 127-134. |
| [9] | 高鹏飞, 席飞虎, 张泽宇, 胡凯强, 陈凯, 魏文桃, 丁家治, 顾连峰. 植物VIGS技术及其在林业科学中的研究进展[J]. 生物技术通报, 2021, 37(5): 141-153. |
| [10] | 王芳, 孙立娇, 赵晓宇, 王婕婉, 宋兴舜. 植物NAC转录因子的研究进展[J]. 生物技术通报, 2019, 35(4): 88-93. |
| [11] | 袁义杭, 张鹤华, 游韩莉, 张凌云. 青杄PwNAC42基因的克隆及表达模式分析[J]. 生物技术通报, 2018, 34(3): 113-120. |
| [12] | 王春雨, 张茜. 植物NAC转录因子功能研究进展[J]. 生物技术通报, 2018, 34(11): 8-14. |
| [13] | 孙威,许奕,许桂莺,孙佩光,宋顺,常胜合. 病毒诱导的基因沉默及其在植物研究中的应用[J]. 生物技术通报, 2015, 31(10): 105-110. |
| [14] | 康桂娟;曾日中;聂智毅;黎瑜;代龙军;段翠芳;. 植物NAC转录因子的研究进展[J]. , 2012, 0(11): 21-26. |
| [15] | 高世庆;王永波;唐益苗;徐蓓;马锦绣;陈京瑞;柳珊;张风廷;赵昌平;. 长穗偃麦草EeNAC9基因功能初步研究[J]. , 2011, 0(06): 47-52. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||