








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (1): 157-167.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0129
Previous Articles Next Articles
CEN Xiao-long1( ), LEI Xi1, MA Shi-yun1, CHEN Qian-ru1, HUANG Zun-xi1,2,3,4, ZHOU Jun-pei1,2,3,4, ZHANG Rui1,2,3,4(
), LEI Xi1, MA Shi-yun1, CHEN Qian-ru1, HUANG Zun-xi1,2,3,4, ZHOU Jun-pei1,2,3,4, ZHANG Rui1,2,3,4( )
)
Received:2021-02-02
Online:2022-01-26
Published:2022-02-22
Contact:
ZHANG Rui
E-mail:1248617002@qq.com;sharezr@126.com
CEN Xiao-long, LEI Xi, MA Shi-yun, CHEN Qian-ru, HUANG Zun-xi, ZHOU Jun-pei, ZHANG Rui. Heterologous Expression and Characterization of the Hyaluronic Acid Lyase HylS from Staphylococcus aureus[J]. Biotechnology Bulletin, 2022, 38(1): 157-167.
| 透明质酸裂解酶 Hyaluronic acid lyase | HylS | HAase | - | HAase-B | HCLaseM | HCLase |
|---|---|---|---|---|---|---|
| 序列一致性a Sequence identity a | 100 | 27.4 | 24.1 | 22.4 | 20.7 | 19.6 |
| 带电荷氨基酸残基 Charged amino acids(RKHDE) | 31.0 | 26.0 | 25.5 | 24.6 | 21.5 | 21.6 |
| 酸性氨基酸残基 Acidic amino acids(DE) | 14.7 | 12.4 | 11.6 | 13.2 | 10.5 | 9.5 |
| 碱性氨基酸残基 Basic amino acids(KR) | 14.4 | 11.3 | 12.0 | 10.1 | 8.7 | 9.4 |
| 极性氨基酸残基 Polar amino acids(NCQSTY) | 32.5 | 28.7 | 36.0 | 29.8 | 24.0 | 28.6 |
| 疏水氨基酸残基 Hydrophobic amino acids(AILFWV) | 26.4 | 35.4 | 28.9 | 32.4 | 39.2 | 35.8 |
| D Asp | 8.8 | 6.2 | 6.4 | 6.3 | 5.9 | 5.8 |
| E Glu | 5.9 | 6.3 | 5.1 | 6.9 | 4.6 | 3.7 |
| C Cys | 0.2 | 0.5 | 0 | 0.1 | 0.6 | 0.3 |
| Y Tyr | 4.6 | 4.3 | 4.0 | 3.9 | 1.9 | 4.0 |
| H His | 1.9 | 2.3 | 1.9 | 1.3 | 2.4 | 2.7 |
| K Lys | 12.4 | 7.8 | 9.2 | 6.2 | 1.0 | 4.4 |
| R Arg | 2.0 | 3.5 | 2.8 | 3.9 | 7.7 | 4.9 |
| 蛋白序列登陆号Protein accession numbers in GenBank database | AYU99970 | AKM20831 | CAD46929 | AHB61202 | QGL52623 | AIL54323 |
| 来源Source | Staphylococcus aureus STA | Streptococcus zooe-pidemicus MF002 | Streptococcus aga- lactiae NEM316 | Bacillus sp. A50 | Microbacterium sp. H14 | Vibrio sp. FC509 |
Table 1 Amino acid residues frequencies of hyaluronic acid(HA)lyases in PL8 family
| 透明质酸裂解酶 Hyaluronic acid lyase | HylS | HAase | - | HAase-B | HCLaseM | HCLase |
|---|---|---|---|---|---|---|
| 序列一致性a Sequence identity a | 100 | 27.4 | 24.1 | 22.4 | 20.7 | 19.6 |
| 带电荷氨基酸残基 Charged amino acids(RKHDE) | 31.0 | 26.0 | 25.5 | 24.6 | 21.5 | 21.6 |
| 酸性氨基酸残基 Acidic amino acids(DE) | 14.7 | 12.4 | 11.6 | 13.2 | 10.5 | 9.5 |
| 碱性氨基酸残基 Basic amino acids(KR) | 14.4 | 11.3 | 12.0 | 10.1 | 8.7 | 9.4 |
| 极性氨基酸残基 Polar amino acids(NCQSTY) | 32.5 | 28.7 | 36.0 | 29.8 | 24.0 | 28.6 |
| 疏水氨基酸残基 Hydrophobic amino acids(AILFWV) | 26.4 | 35.4 | 28.9 | 32.4 | 39.2 | 35.8 |
| D Asp | 8.8 | 6.2 | 6.4 | 6.3 | 5.9 | 5.8 |
| E Glu | 5.9 | 6.3 | 5.1 | 6.9 | 4.6 | 3.7 |
| C Cys | 0.2 | 0.5 | 0 | 0.1 | 0.6 | 0.3 |
| Y Tyr | 4.6 | 4.3 | 4.0 | 3.9 | 1.9 | 4.0 |
| H His | 1.9 | 2.3 | 1.9 | 1.3 | 2.4 | 2.7 |
| K Lys | 12.4 | 7.8 | 9.2 | 6.2 | 1.0 | 4.4 |
| R Arg | 2.0 | 3.5 | 2.8 | 3.9 | 7.7 | 4.9 |
| 蛋白序列登陆号Protein accession numbers in GenBank database | AYU99970 | AKM20831 | CAD46929 | AHB61202 | QGL52623 | AIL54323 |
| 来源Source | Staphylococcus aureus STA | Streptococcus zooe-pidemicus MF002 | Streptococcus aga- lactiae NEM316 | Bacillus sp. A50 | Microbacterium sp. H14 | Vibrio sp. FC509 |

Fig. 1 Partial amino acid sequences alignment of HylS and PL8 HA lyases Sequences are named as the protein accession numbers in GenBank database. AAK99090 from Streptomyces coelicolor A3(2)(PDB ID:2WCO,2WDA and 2X03). AHB61202 from Bacillus sp. A50. AAK74491 from Streptococcus pneumoniae TIGR4. CAD46929 from S. agalactiae NEM316(PDB ID:1F1S,1I8Q and 1LXM). QGL52623 from Microbacterium sp. H14. Identical and similar amino acids are shaded in black and framed,respectively. The catalytic group is marked using “*”. The hydrophobic patch in hyaluronic acid of bound substrate is marked using “#”

Fig. 2 SDS-PAGE analysis of the purified rHylS M:Protein markers. 1:Cell lysate of uninduced transformant. 2:Cell lysate of induced transformant. 3:Purified recombinant HylS
| 试剂 Reagent | 相对酶活力 Relative activity /% | 试剂 Reagent | 相对酶活力 Relative activity/% | |
|---|---|---|---|---|
| None | 100.0±1.4 | CaCl2 | 60.7±2.0 | |
| MnSO4 | 178.3±1.6 | NiSO4 | 40.7±0.9 | |
| KCl | 123.1±2.8 | PbAc | 39.8±0.9 | |
| FeSO4 | 103.8±2.3 | ZnSO4 | 29.8±0.5 | |
| NaCl | 99.9±0.8 | AlCl3 | 0 | |
| MgSO4 | 97.9±1.1 | β-Mercaptoethanol | 162.7±1.8 | |
| LiCl | 93.8±2.4 | SDS | 91.7±2.1 | |
| CuSO4 | 86.5±1.5 | CTAB | 65.2±1.0 | |
| CoCl2 | 77.4±1.0 | EDTA | 42.8±0.7 |
Table 2 Effects of metal ions and chemical reage-nts on the purified rHylS
| 试剂 Reagent | 相对酶活力 Relative activity /% | 试剂 Reagent | 相对酶活力 Relative activity/% | |
|---|---|---|---|---|
| None | 100.0±1.4 | CaCl2 | 60.7±2.0 | |
| MnSO4 | 178.3±1.6 | NiSO4 | 40.7±0.9 | |
| KCl | 123.1±2.8 | PbAc | 39.8±0.9 | |
| FeSO4 | 103.8±2.3 | ZnSO4 | 29.8±0.5 | |
| NaCl | 99.9±0.8 | AlCl3 | 0 | |
| MgSO4 | 97.9±1.1 | β-Mercaptoethanol | 162.7±1.8 | |
| LiCl | 93.8±2.4 | SDS | 91.7±2.1 | |
| CuSO4 | 86.5±1.5 | CTAB | 65.2±1.0 | |
| CoCl2 | 77.4±1.0 | EDTA | 42.8±0.7 |
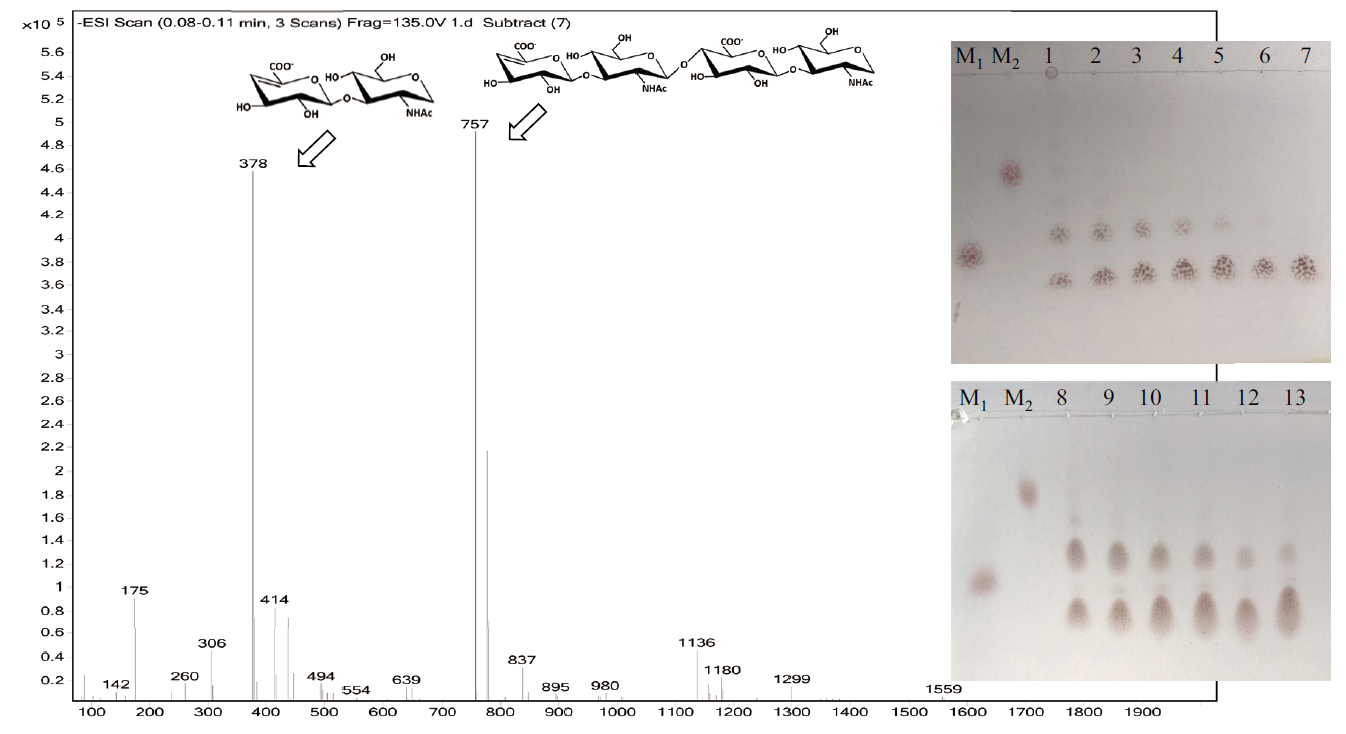
Fig. 5 TLC and ESI-MS analysis of the degraded products of HA by rHylS TLC:M1 and M2 are chitobiose and chitotetraose,respectively. 1-7:rHylS degraded 2% HA for 0.5,1,2,4,6,8 and 10 h,respectively. 8-13:rHylS degraded 5% HA for 0.5,2,4,6,8 and 10 h,respectively. ESI-MS:rHylS degraded 2% HA for 2 h

Fig. 6 Antioxidant activity of the degraded products of HA by rHylS The scavenging ability of the degraded products by rHylS to each free radical is labeled individually according to the least significant difference test among values(P<0.01),and different letters refer to significant differences
| 透明质酸裂解酶 HA lyase | HylS | HAase | - | HAase-B | HCLaseM | HCLase |
|---|---|---|---|---|---|---|
| 最适pH Optimal pH | 5.0 | 6.0 | 6.3 | 6.5 | 7.0 | 8.0 |
| pH稳定性a pH stabilitya | 100%/ pH 5.0/60 min | 95% pH 5.0/60 min | - | 75% pH 5.0/60 min | 70% pH 5.0/12 h | - |
| 最适温度 Optimal temperature/℃ | 45 | 37 | 40 | 44 | 35 | 30 |
| 热稳定性b Thermostabilityb | 60%/50℃/60 min | 70%/50℃/60 min | - | 50%/50℃/10 min | 50%/40℃/60 min | 50%/40℃/60 min |
| 激活剂 Activators | Mn2+/K+ β-Mercaptoethanol | Ca2+/Mg2+/Co2+ | Mg2+ | Ca2+/Mg2+/Ni2+ | - | Li+/Na+/K+ |
| 抑制剂 Inhibitors | Ni2+/Zn2+/Al3+/Pb2+ EDTA | Zn2+/Cu2+ | Zn2+/Al3+/Cu2+ /Fe2+/Mn2+ | Zn2+/ Cu2+/ SDS/EDTA | Hg2+/SDS | Ag+/Co2+/Hg2+/ Ni2+/Cu2+/Zn2+/Fe3+/Cr3+ |
| 底物特异性c Substrate specificityc | HA | - | HA/CS/DS | HA/CS | HA/CS/DS | HA/CS |
| 比活d Specific activity / (U·mg-1)d | 1.6×105 | 10.5 | 8.1×104 | 1.0×106 | 278.3 | 4.5×105 |
| 检测方法e Measurement methode | 紫外法 UV spectrophotometry | 二硝基水杨酸比色法 DNS colorimetry | CATB浊度法 CATB turbidimetry | BSA浊度法 BSA turbidimetry | 紫外法 UV spectroph-otometry | 紫外法 UV spectrophotometry |
| 降解模式 Degrading pattern | 内切 Endo | - | 外切 Exo | 内切 Endo | 内切 Endo | 内切 Endo |
| 来源 Source | S. aureus STA | S. zooepidemicus MF002 | S. agalactiae NEM316 | Bacillus sp. A50 | Microbacterium sp.H14 | Vibrio sp. FC509 |
| 参考文献References | This study | [17] | [18-19] | [20] | [21] | [22] |
Table 3 Characteristics of HA lyases in PL8 family
| 透明质酸裂解酶 HA lyase | HylS | HAase | - | HAase-B | HCLaseM | HCLase |
|---|---|---|---|---|---|---|
| 最适pH Optimal pH | 5.0 | 6.0 | 6.3 | 6.5 | 7.0 | 8.0 |
| pH稳定性a pH stabilitya | 100%/ pH 5.0/60 min | 95% pH 5.0/60 min | - | 75% pH 5.0/60 min | 70% pH 5.0/12 h | - |
| 最适温度 Optimal temperature/℃ | 45 | 37 | 40 | 44 | 35 | 30 |
| 热稳定性b Thermostabilityb | 60%/50℃/60 min | 70%/50℃/60 min | - | 50%/50℃/10 min | 50%/40℃/60 min | 50%/40℃/60 min |
| 激活剂 Activators | Mn2+/K+ β-Mercaptoethanol | Ca2+/Mg2+/Co2+ | Mg2+ | Ca2+/Mg2+/Ni2+ | - | Li+/Na+/K+ |
| 抑制剂 Inhibitors | Ni2+/Zn2+/Al3+/Pb2+ EDTA | Zn2+/Cu2+ | Zn2+/Al3+/Cu2+ /Fe2+/Mn2+ | Zn2+/ Cu2+/ SDS/EDTA | Hg2+/SDS | Ag+/Co2+/Hg2+/ Ni2+/Cu2+/Zn2+/Fe3+/Cr3+ |
| 底物特异性c Substrate specificityc | HA | - | HA/CS/DS | HA/CS | HA/CS/DS | HA/CS |
| 比活d Specific activity / (U·mg-1)d | 1.6×105 | 10.5 | 8.1×104 | 1.0×106 | 278.3 | 4.5×105 |
| 检测方法e Measurement methode | 紫外法 UV spectrophotometry | 二硝基水杨酸比色法 DNS colorimetry | CATB浊度法 CATB turbidimetry | BSA浊度法 BSA turbidimetry | 紫外法 UV spectroph-otometry | 紫外法 UV spectrophotometry |
| 降解模式 Degrading pattern | 内切 Endo | - | 外切 Exo | 内切 Endo | 内切 Endo | 内切 Endo |
| 来源 Source | S. aureus STA | S. zooepidemicus MF002 | S. agalactiae NEM316 | Bacillus sp. A50 | Microbacterium sp.H14 | Vibrio sp. FC509 |
| 参考文献References | This study | [17] | [18-19] | [20] | [21] | [22] |
| [1] |
Tolg C, Telmer P, Turley E. Specific sizes of hyaluronan oligosaccharides stimulate fibroblast migration and excisional wound repair[J]. PLoS One, 2014, 9(2):e88479.
doi: 10.1371/journal.pone.0088479 URL |
| [2] | 李憬昱, 王凤山. 低分子质量和寡聚透明质酸制备、活性与应用研究进展[J]. 药物生物技术, 2019, 26(1):64-68. |
| Li JY, Wang FS. Research progress in the preparation methods, bioactivities and applications of low molecular weight and oligomeric hyaluronic acid[J]. Pharm Biotechnol, 2019, 26(1):64-68. | |
| [3] | 雷曦, 张蕊, 黄遵锡, 等. 透明质酸酶的研究进展[J]. 微生物学通报, 2021, 48(3):882-895. |
| Lei X, Zhang R, Huang ZX, et al. Research progress of hyaluronidases[J]. Microbiol China, 2021, 48(3):882-895. | |
| [4] | Wang W, Wang J, Li F. Hyaluronidase and chondroitinase[J]. Adv Exp Med Biol, 2017, 925:75-87. |
| [5] |
Duran-Reynals F. Tissue permeability and the spreading factors in infection 1[J]. Bacteriol Rev, 1942, 6(4):197-252.
doi: 10.1128/br.6.4.197-252.1942 pmid: 16350083 |
| [6] |
El-Safory NS, Fazary AE, Lee CK. Hyaluronidases, a group of glycosidases:Current and future perspectives[J]. Carbohydr Polym, 2010, 81(2):165-181.
doi: 10.1016/j.carbpol.2010.02.047 URL |
| [7] | Rungsa P, Janpan P, Saengkun Y, et al. Heterologous expression and mutagenesis of recombinant Vespa affinis hyaluronidase protein(rVesA2)[J]. J Venom Animals Toxins Trop Dis, 2019, 25:e20190030. |
| [8] |
Bordon KC, Wiezel GA, Amorim FG, et al. Arthropod venom Hyaluronidases:biochemical properties and potential applications in medicine and biotechnology[J]. J Venom Anim Toxins Incl Trop Dis, 2015, 21:43.
doi: 10.1186/s40409-015-0042-7 pmid: 26500679 |
| [9] |
Huang H, Liang Q, Wang Y, et al. High-level constitutive expression of leech hyaluronidase with combined strategies in recombinant Pichia pastoris[J]. Appl Microbiol Biotechnol, 2020, 104(4):1621-1632.
doi: 10.1007/s00253-019-10282-7 pmid: 31907577 |
| [10] |
Garron ML, Cygler M. Structural and mechanistic classification of uronic acid-containing polysaccharide lyases[J]. Glycobiology, 2010, 20(12):1547-1573.
doi: 10.1093/glycob/cwq122 URL |
| [11] |
Viborg AH, Terrapon N, Lombard V, et al. A subfamily roadmap of the evolutionarily diverse glycoside hydrolase family 16(GH16)[J]. J Biol Chem, 2019, 294(44):15973-15986.
doi: 10.1074/jbc.RA119.010619 URL |
| [12] | 张海洋. 透明质酸酶的表达及酶法制备低聚透明质酸[D]. 天津:天津科技大学, 2016. |
| Zhang HY. Expression of A hyalronidase and preparation oligomers hyaluronate by enzymolysis[D]. Tianjin:Tianjin University of Science & Technology, 2016. | |
| [13] | 李帅帅. 一种节杆菌来源的透明质酸酶的研究[D]. 济南:山东大学, 2017. |
| Li SS. Study on a hyaluronidase from Arthrobacter sp[D]. Jinan:Shandong University, 2017. | |
| [14] |
Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server[J]. Nucleic Acids Res, 2014, 42(W1):W320-W324.
doi: 10.1093/nar/gku316 URL |
| [15] |
Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL:homology modelling of protein structures and complexes[J]. Nucleic Acids Res, 2018, 46(w1):W296-W303.
doi: 10.1093/nar/gky427 URL |
| [16] |
Zhu YP, Li XT, Sun BG, et al. Properties of an alkaline-tolerant, thermostable xylanase from Streptomyces chartreusis L1105, suitable for xylooligosaccharide production[J]. J Food Sci, 2012, 77(5):C506-C511.
doi: 10.1111/jfds.2012.77.issue-5 URL |
| [17] | 陈怡斐. 重组透明质酸酶的表达、纯化及其酶学性质研究[D]. 上海:上海应用技术学院, 2016. |
| Chen YF. Expression, purification and enzymatic proporties of recombinant hyaluronidase[D]. Shanghai:Shanghai Institute of Technology, 2016. | |
| [18] |
Ozegowski JH, Günther E, Reichardt W. Purification and characterization of hyaluronidase from Streptococcus agalactiae[J]. Zentralbl Bakteriol, 1994, 280(4):497-506.
doi: 10.1016/S0934-8840(11)80509-8 URL |
| [19] |
Li S, Jedrzejas MJ. Hyaluronan binding and degradation by Streptococcus agalactiae hyaluronate lyase[J]. J Biol Chem, 2001, 276(44):41407-41416.
pmid: 11527972 |
| [20] |
Guo X, Shi Y, Sheng J, et al. A novel hyaluronidase produced by Bacillus sp. A50[J]. PLoS One, 2014, 9(4):e94156.
doi: 10.1371/journal.pone.0094156 URL |
| [21] |
Sun JH, Han X, Song GR, et al. Cloning, expression, and characterization of a new glycosaminoglycan lyase from Microbacterium sp. H14[J]. Mar Drugs, 2019, 17(12):681.
doi: 10.3390/md17120681 URL |
| [22] |
Han W, Wang W, Zhao M, et al. A novel eliminase from a marine bacterium that degrades hyaluronan and chondroitin sulfate[J]. J Biol Chem, 2014, 289(40):27886-27898.
doi: 10.1074/jbc.M114.590752 URL |
| [23] |
Zhao YJ, Zhang YH, Cao Y, et al. Structural analysis of alkaline β-mannanase from alkaliphilic Bacillus sp. N16-5:implications for adaptation to alkaline conditions[J]. PLoS One, 2011, 6(1):e14608. DOI: 10.1371/journal.pone.0014608.
doi: 10.1371/journal.pone.0014608 URL |
| [24] |
Liu Q, Yang PL, Luo HY, et al. A novel endo-1, 4-β-mannanase from Bispora antennata with good adaptation and stability over a broad pH range[J]. Appl Biochem Biotechnol, 2012, 166(6):1442-1453.
doi: 10.1007/s12010-011-9537-z URL |
| [25] |
Nukui M, Taylor KB, McPherson DT, et al. The function of hydrophobic residues in the catalytic cleft of Streptococcus pneumoniae hyaluronate lyase. Kinetic characterization of mutant enzyme forms[J]. J Biol Chem, 2003, 278(5):3079-3088.
doi: 10.1074/jbc.M204999200 URL |
| [26] |
Jedrzejas MJ, Mello LV, de Groot BL, et al. Mechanism of hyaluronan degradation by Streptococcus pneumoniae hyaluronate lyase. Structures of complexes with the substrate[J]. J Biol Chem, 2002, 277(31):28287-28297.
pmid: 11991948 |
| [27] | 张晓娟, 宫倩红. 特异性透明质酸酶的研究进展[J]. 青岛大学学报:医学版, 2018, 54(6):750-752, 755. |
| Zhang XJ, Gong QH. Research progress of specific hyaluronidases[J]. J Qingdao Univ:Med Sci, 2018, 54(6):750-752, 755. | |
| [28] | 王晓宇, 杜国荣, 李华. 抗氧化能力的体外测定方法研究进展[J]. 食品与生物技术学报, 2012, 31(3):247-252. |
| Wang XY, Du GR, Li H. Progress of analytical methods for antioxidant capacity in vitro[J]. J Food Sci Biotechnol, 2012, 31(3):247-252. | |
| [29] | 柯春林, 乔德亮, 曾晓雄. 低分子量透明质酸的制备及其抗氧化活性的研究[J]. 食品工业科技, 2010, 31(1):107-111. |
| Ke CL, Qiao DL, Zeng XX. Preparation of the low molecular weight hyaluronic acid and its antioxidant activity[J]. Sci Technol Food Ind, 2010, 31(1):107-111. | |
| [30] | 李密, 李和生, 张丽媛, 等. 乌贼眼透明质酸的体外抗氧化性及对小鼠创愈性质的研究[J]. 中国食品学报, 2017, 17(10):30-36. |
| Li M, Li HS, Zhang LY, et al. Studies on the antioxidant and wound heal properties of hyaluronic acid from cuttlefish[J]. J Chin Inst Food Sci Technol, 2017, 17(10):30-36. | |
| [31] | 高瑞昌, 陈辉, 李来好, 等. 低分子量罗非鱼眼透明质酸的制备及其抗氧化性研究[J]. 食品工业科技, 2015, 36(3):60-64. |
| Gao RC, Chen H, Li LH, et al. Preparation and antioxidant properties of low molecular weigh hyaluronic acid from tilapia eye[J]. Sci Technol Food Ind, 2015, 36(3):60-64. |
| [1] | KANG Ling-yun, HAN Lu-lu, HAN De-ping, CHEN Jian-sheng, GAN Han-ling, XING Kai, MA You-ji, CUI Kai. Effect of Melatonin on Protecting the Jejunum Mucosal Epithelial Cells from Oxidative Stress Damage [J]. Biotechnology Bulletin, 2023, 39(9): 291-299. |
| [2] | YOU Zi-juan, CHEN Han-lin, DENG Fu-cai. Research Progress in the Extraction and Functional Activities of Bioactive Peptides from Fish Skin [J]. Biotechnology Bulletin, 2023, 39(7): 91-104. |
| [3] | WANG Chun-yu, LI Zheng-jun, WANG Ping, ZHANG Li-xia. Physiological and Biochemical Analysis of Drought Resistance in Sorghum Cuticular Wax-deficient Mutant sb1 [J]. Biotechnology Bulletin, 2023, 39(5): 160-167. |
| [4] | CHEN Xiao-meng, ZHANG Xue-jing, ZHANG Huan, ZHANG Bao-jiang, SU Yan. Prokaryotic Expression of Recombinant Bovine Mastitis Staphylococcus aureus GapC Protein and Identification of Its B-cell Epitopes [J]. Biotechnology Bulletin, 2023, 39(5): 306-313. |
| [5] | YANG Mao, LIN Yu-feng, DAI Yang-shuo, PAN Su-jun, PENG Wei-ye, YAN Ming-xiong, LI Wei, WANG Bing, DAI Liang-ying. OsDIS1 Negatively Regulates Rice Drought Tolerance Through Antioxidant Pathways [J]. Biotechnology Bulletin, 2023, 39(2): 88-95. |
| [6] | ZHAO Jia, ZHAO Fei-yan, SHEN Xin, GAO Guang-qi, SUN Zhi-hong. Advances in the Antioxidant Activities of Lactic Acid Bacteria and Their Applications [J]. Biotechnology Bulletin, 2023, 39(11): 182-190. |
| [7] | ZHU Jin-cheng, YANG Yang, LOU Hui, ZHANG Wei. Regulation of Fusarium wilt Resistance in Cotton by Exogenous Melatonin [J]. Biotechnology Bulletin, 2023, 39(1): 243-252. |
| [8] | CHENG Shen-wei, ZHANG Ke-qiang, LIANG Jun-feng, LIU Fu-yuan, GAO Xing-liang, DU Lian-zhu. Establishment of a Triple Droplet Digital PCR Quantitative Detection Method for Typical Pathogenic Bacteria in Livestock and Poultry Manure [J]. Biotechnology Bulletin, 2022, 38(9): 271-280. |
| [9] | MAO Guo-tao, WANG Jie, WANG Kai, WANG Fang-yuan, CAO Le-yan, ZHANG Hong-sen, SONG An-dong. Characterization of Laccase TaLac from Thermus aquaticus and Its Application in Removing Malachite Green Dye [J]. Biotechnology Bulletin, 2022, 38(4): 261-268. |
| [10] | CHANG Qing, SHU Yue-rong, WANG Wen-tao, JIANG Hao, YAN Quan-de, QIAN Zheng, GAO Xue-chun, WU Jin-hong, ZHANG Yong. Heterologous Expression and Characterization of Endo-type Alginate Lyase from Yeosuana marina sp. JLT21 [J]. Biotechnology Bulletin, 2022, 38(2): 123-131. |
| [11] | ZHANG Feng-wen, ZHOU Li-ya, DONG Chao, SHI Yan-mao. Purification of Antioxidant Peptides from Natto Supernatant and Study on Its Activity [J]. Biotechnology Bulletin, 2022, 38(2): 158-165. |
| [12] | MA Yan-qin, QIU Yi-bin, LI Sha, XU Hong. Research Progress in the Biosynthesis and Metabolic Engineering of Hyaluronic Acid [J]. Biotechnology Bulletin, 2022, 38(2): 252-262. |
| [13] | SUN Bao-ting, QIU Meng-xia, WANG Zi-chen, WANG Zi-yuan, CUI Jian-dong, JIA Shi-ru. Preparation of @ZIF-8 Immobilized Enzyme by Using Cysteine as Auxiliary Reagent and Its Characterization [J]. Biotechnology Bulletin, 2021, 37(8): 221-232. |
| [14] | HAO Xiang-yang, LIU Fan, WU Huan, WANG Bin, SUN Xue-li, XIANG Lei-lei, WANG Tian-chi, LAI Zhong-xiong, CHENG Chun-zhen. Cloning and Expression Analysis of GjPAL Genes in Gerbera jamesonni [J]. Biotechnology Bulletin, 2021, 37(6): 13-23. |
| [15] | XU Kun, YANG Ai-jiang, HU Xia, ZOU Hai-tao, LI Bin, LIU Ji. Antimony Accumulation and Its Effect on Antioxidation System in Different Tissues of Danio rerio [J]. Biotechnology Bulletin, 2021, 37(4): 145-154. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||