








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (12): 227-234.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0616
Previous Articles Next Articles
WEI Wen-qing( ), XIE Ze-xiong(
), XIE Ze-xiong( )
)
Received:2021-05-12
Online:2021-12-26
Published:2022-01-19
Contact:
XIE Ze-xiong
E-mail:wqwei@tju.edu.cn;xzx@tju.edu.cn
WEI Wen-qing, XIE Ze-xiong. Mechanism of Abnormal Chromosomal Segregation in Yeast Mitosis[J]. Biotechnology Bulletin, 2021, 37(12): 227-234.
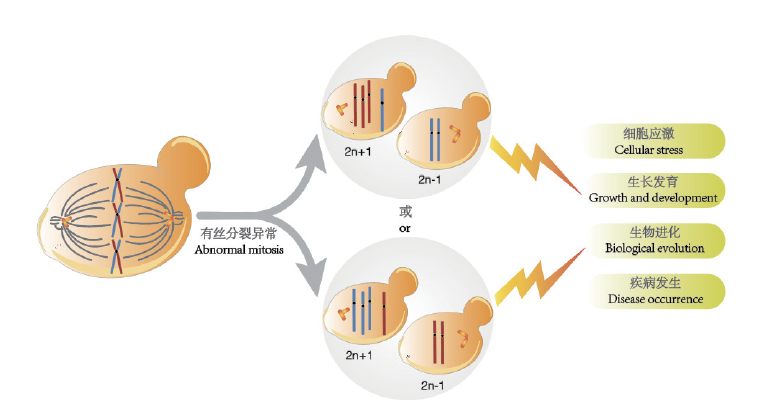
Fig.1 Formation and influence of yeast aneuploidy Abnormal mitosis process in yeast cells will lead to the formation of aneuploidy,which will affect cell stress,growth and development,biological evolution and the occurrence of diseases
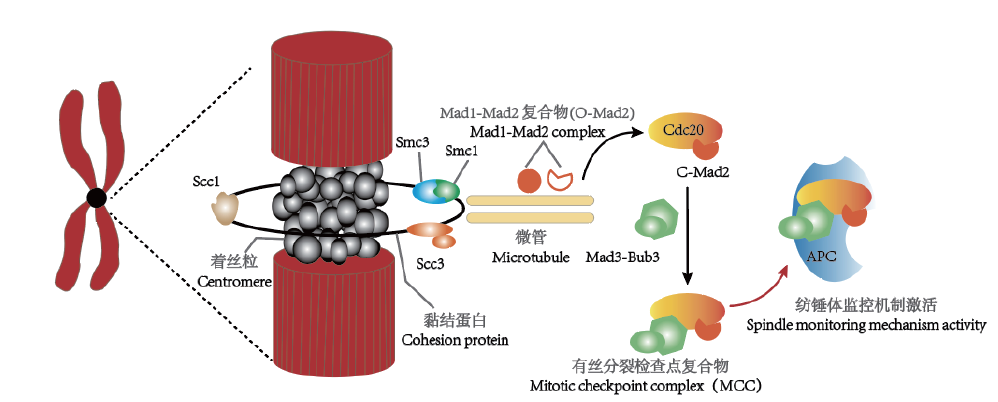
Fig.2 Sister chromatid adhesion protein and spindle monitoring mechanism Smc1,Smc3,Rad21 and SA/Stg2 together form a chromatid adhesive protein complex,forming a single ring model around the centromere,capturing sister chromatid to regulate polymerization. If the kinetochore-microtubule combination is not correct in the pre-middle stage of mitosis,the mitotic checkpoint complex(MCC)binds to the advanced-stage complex(APC)to activate the spindle assembly monitoring mechanism to block the process of cell mitosis
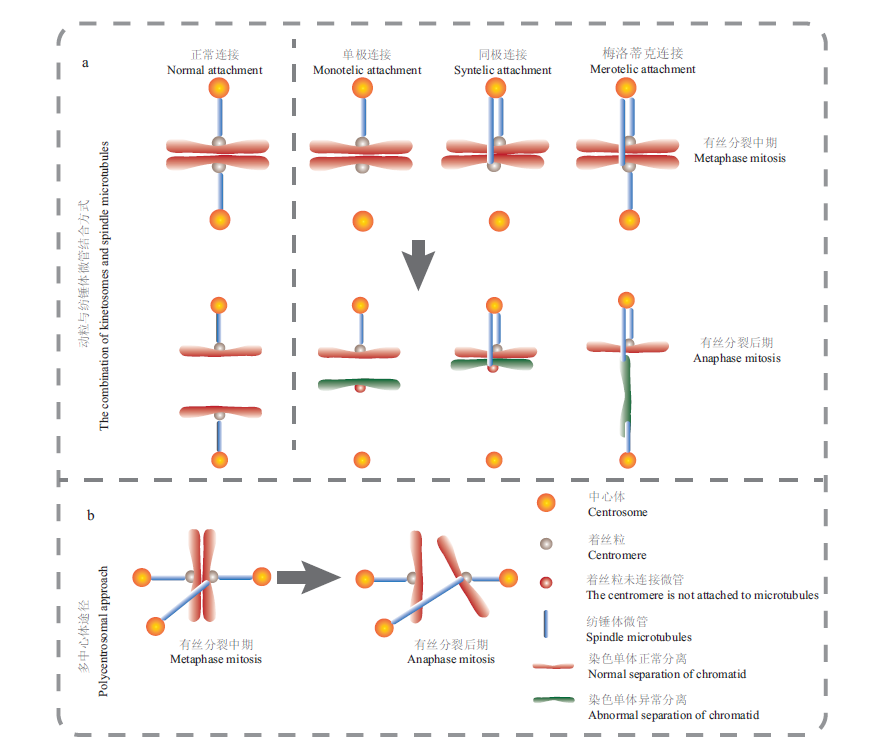
Fig.3 Miscellaneous kinetochore-microtubule binding and polycentric pathway a:During mitosis,the kinetochore-microtubules are combined in different ways;the bipolar connection is the kinetochore-microtubule connection that the cell completes the normal mitosis process;the Monotic connection,Syntelic connection and Merotelic connection will cause abnormal chromosome segregation and lead to the production of aneuploid yeast cells. b:When there are more than two centrosomes in a cell,centrosomes in different directions are combined with the kinetochore through microtubules,and the chromosomes will be unevenly separated due to uneven force to form aneuploid cells
| [1] |
Todd RT, Forche A, Selmecki A. Ploidy variation in fungi:polyploidy, aneuploidy, and genome evolution[J]. Microbiol Spectr, 2017, 5(4). DOI: 10.1128/microbiolspec.funk-0051-2016.
doi: 10.1128/microbiolspec.funk-0051-2016 |
| [2] |
Weaver BA, Cleveland DW. Does aneuploidy cause cancer?[J]. Curr Opin Cell Biol, 2006, 18(6):658-667.
pmid: 17046232 |
| [3] |
Ohbayashi T, Oikawa K, Yamada K, et al. Unscheduled overexpression of human WAPL promotes chromosomal instability[J]. Biochem Biophys Res Commun, 2007, 356(3):699-704.
doi: 10.1016/j.bbrc.2007.03.037 URL |
| [4] |
Xie ZX, Li BZ, Mitchell LA, et al. “Perfect” designer chromosome V and behavior of a ring derivative[J]. Science, 2017, 355(6329):eaaf4704.
doi: 10.1126/science.aaf4704 URL |
| [5] |
Selmecki AM, Maruvka YE, Richmond PA, et al. Polyploidy can drive rapid adaptation in yeast[J]. Nature, 2015, 519(7543):349-352.
doi: 10.1038/nature14187 URL |
| [6] |
Deutschbauer AM, Jaramillo DF, Proctor M, et al. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast[J]. Genetics, 2005, 169(4):1915-1925.
pmid: 15716499 |
| [7] |
Torres EM, Williams BR, Amon A. Aneuploidy:cells losing their balance[J]. Genetics, 2008, 179(2):737-746.
doi: 10.1534/genetics.108.090878 URL |
| [8] |
Beach RR, Ricci-Tam C, Brennan CM, et al. Aneuploidy causes non-genetic individuality[J]. Cell, 2017, 169(2):229-242.e21.
doi: 10.1016/j.cell.2017.03.021 URL |
| [9] |
Harigaya Y, Yamamoto M. Molecular mechanisms underlying the mitosis-meiosis decision[J]. Chromosome Res, 2007, 15(5):523-537.
pmid: 17674143 |
| [10] |
Yamamoto M. Regulation of meiosis in fission yeast[J]. Cell Struct Funct, 1996, 21(5):431-436.
pmid: 9118252 |
| [11] | 吕磊. 卵巢癌非整倍体细胞发生途径及其机制的研究[D]. 合肥:中国科学技术大学, 2011. |
| Lv L. The pathway and mechanism of aneuploidy generation during ovarian cancer neoplastic progression[D]. Hefei:University of Science and Technology of China, 2011. | |
| [12] |
Thompson SL, Bakhoum SF, Compton DA. Mechanisms of chromosomal instability[J]. Curr Biol, 2010, 20(6):R285-R295.
doi: 10.1016/j.cub.2010.01.034 URL |
| [13] | Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time[J]. Nat Rev Mol Cell Biol, 2007, 8(5):379-393. |
| [14] |
Dorsett D. Cohesin:genomic insights into controlling gene transcription and development[J]. Curr Opin Genet Dev, 2011, 21(2):199-206.
doi: 10.1016/j.gde.2011.01.018 pmid: 21324671 |
| [15] |
Indjeian VB, Murray AW. Budding yeast mitotic chromosomes have an intrinsic bias to biorient on the spindle[J]. Curr Biol, 2007, 17(21):1837-1846.
pmid: 17980598 |
| [16] | Mehta GD, Rizvi SM, Ghosh SK. Cohesin:a guardian of genome integrity[J]. Biochim Biophys Acta, 2012, 1823(8):1324-1342. |
| [17] |
Losada A, Hirano T. Dynamic molecular linkers of the genome:the first decade of SMC proteins[J]. Genes Dev, 2005, 19(11):1269-1287.
doi: 10.1101/gad.1320505 URL |
| [18] |
Ivanov D, Schleiffer A, Eisenhaber F, et al. Eco1 is a novel acetyltransferase that can acetylate proteins involved in cohesion[J]. Curr Biol, 2002, 12(4):323-328.
doi: 10.1016/S0960-9822(02)00681-4 URL |
| [19] |
Zhang J, Shi X, Li Y, et al. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast[J]. Mol Cell, 2008, 31(1):143-151.
doi: 10.1016/j.molcel.2008.06.006 URL |
| [20] | Huang CE, Milutinovich M, Koshland D. Rings, bracelet or snaps:fashionable alternatives for Smc complexes[J]. Philos Trans Royal Soc Lond Ser B Biol Sci, 2005, 360(1455):537-542. |
| [21] |
Zhang N, Kuznetsov SG, Sharan SK, et al. A handcuff model for the cohesin complex[J]. J Cell Biol, 2008, 183(6):1019-1031.
doi: 10.1083/jcb.200801157 URL |
| [22] |
Kim N. How might cohesin hold sister chromatids together?[J]. Phil Trans R Soc B, 2005, 360(1455):483-496.
doi: 10.1098/rstb.2004.1604 URL |
| [23] |
Surcel A, Koshland D, Ma H, et al. Cohesin interaction with centromeric minichromosomes shows a multi-complex rod-shaped structure[J]. PLoS One, 2008, 3(6):e2453.
doi: 10.1371/journal.pone.0002453 URL |
| [24] |
Allshire RC. Centromeres, checkpoints and chromatid cohesion[J]. Curr Opin Genet Dev, 1997, 7(2):264-273.
pmid: 9115433 |
| [25] |
Li R, Murray AW. Feedback control of mitosis in budding yeast[J]. Cell, 1991, 66(3):519-531.
pmid: 1651172 |
| [26] |
May KM, Paldi F, Hardwick KG. Fission yeast Apc15 stabilizes MCC-Cdc20-APC/C complexes, ensuring efficient Cdc20 ubiquitination and checkpoint arrest[J]. Curr Biol, 2017, 27(8):1221-1228.
doi: 10.1016/j.cub.2017.03.013 URL |
| [27] |
Chung E, Chen RH. Spindle checkpoint requires Mad1-bound and Mad1-free Mad2[J]. Mol Biol Cell, 2002, 13(5):1501-1511.
doi: 10.1091/mbc.02-01-0003 URL |
| [28] |
Castro A, Bernis C, Vigneron S, et al. The anaphase-promoting complex:a key factor in the regulation of cell cycle[J]. Oncogene, 2005, 24(3):314-325.
pmid: 15678131 |
| [29] |
Fraschini R, Beretta A, Sironi L, et al. Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores[J]. EMBO J, 2001, 20(23):6648-6659.
pmid: 11726501 |
| [30] |
Yang M, Li B, Liu CJ, et al. Insights into Mad2 regulation in the spindle checkpoint revealed by the crystal structure of the symmetric Mad2 dimer[J]. PLoS Biol, 2008, 6(3):e50.
doi: 10.1371/journal.pbio.0060050 URL |
| [31] | 晁春江. 裂殖酵母中核仁蛋白Dnt1调控后期促进复合物功能的研究[D]. 厦门:厦门大学, 2018. |
| Chao CJ. A study on the regulation of anaphase-promoting complex/cyclosome by nucleolar protein Dnt1 in fission yeast[D]. Xiamen:Xiamen University, 2018. | |
| [32] | Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes[J]. Biochim Biophys Acta, 2004, 1695(1/2/3):189-207. |
| [33] |
Reddy SK, Rape M, Margansky WA, et al. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation[J]. Nature, 2007, 446(7138):921-925.
doi: 10.1038/nature05734 URL |
| [34] |
Kim ST, Xu B, Kastan MB. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage[J]. Genes Dev, 2002, 16(5):560-570.
doi: 10.1101/gad.970602 URL |
| [35] |
Watrin E, Peters JM. The cohesin complex is required for the DNA damage-induced G2/M checkpoint in mammalian cells[J]. EMBO J, 2009, 28(17):2625-2635.
doi: 10.1038/emboj.2009.202 pmid: 19629043 |
| [36] |
Lopes CS, Sunkel CE. The spindle checkpoint:from normal cell division to tumorigenesis[J]. Arch Med Res, 2003, 34(3):155-165.
doi: 10.1016/S0188-4409(03)00024-9 URL |
| [37] |
de Voer RM, Hoogerbrugge N, Kuiper RP. Spindle-assembly checkpoint and gastrointestinal cancer[J]. N Engl J Med, 2011, 364(13):1279-1280.
doi: 10.1056/NEJMc1101053 URL |
| [38] |
Lim HH, Zhang T, Surana U. Regulation of centrosome separation in yeast and vertebrates:common threads[J]. Trends Cell Biol, 2009, 19(7):325-333.
doi: 10.1016/j.tcb.2009.03.008 URL |
| [39] |
Lampson MA, Renduchitala K, Khodjakov A, et al. Correcting improper chromosome-spindle attachments during cell division[J]. Nat Cell Biol, 2004, 6(3):232-237.
doi: 10.1038/ncb1102 URL |
| [40] | 胡逸超, 许雨晨, 鹿宗贵, 等. Dam1复合体在动粒-微管互作中的作用[J]. 基因组学与应用生物学, 2015, 34(4):896-901. |
| Hu YC, Xu YC, Lu ZG, et al. Functions of Dam1 complex in outer kinetochore[J]. Genom Appl Biol, 2015, 34(4):896-901. | |
| [41] |
Liu X, McLeod I, Anderson S, et al. Molecular analysis of kinetochore architecture in fission yeast[J]. EMBO J, 2005, 24(16):2919-2930.
doi: 10.1038/sj.emboj.7600762 URL |
| [42] |
McAinsh AD, Tytell JD, Sorger PK. Structure, function, and regulation of budding yeast kinetochores[J]. Annu Rev Cell Dev Biol, 2003, 19:519-539.
pmid: 14570580 |
| [43] | 梁前进. 着丝粒和动粒[J]. 生物学通报, 2012, 47(4):1-4. |
| Liang QJ. Centromere and kinetochore[J]. Bull Biol, 2012, 47(4):1-4. | |
| [44] |
Taylor SS, Scott MIF, Holland AJ. The spindle checkpoint:a quality control mechanism which ensures accurate chromosome segregation[J]. Chromosom Res, 2004, 12(6):599-616.
doi: 10.1023/B:CHRO.0000036610.78380.51 URL |
| [45] |
Cheeseman IM, Brew C, Wolyniak M, et al. Implication of a novel multiprotein Dam1p complex in outer kinetochore function[J]. J Cell Biol, 2001, 155(7):1137-1145.
pmid: 11756468 |
| [46] |
Meraldi P, Draviam VM, Sorger PK. Timing and checkpoints in the regulation of mitotic progression[J]. Dev Cell, 2004, 7(1):45-60.
doi: 10.1016/j.devcel.2004.06.006 URL |
| [47] | 李强, 吴燕华, 闫晓梅, 等. 极光(aurora)激酶在细胞有丝分裂和肿瘤形成中的重要功能[J]. 生命科学, 2005, 17(5):424-432 |
| Li Q, Wu YH, Yan XM, et al. The key functions of aurora kinases during cell mitosis and tumorigenesis[J]. Chin Bull Life Sci, 2005, 17(5):424-432 | |
| [48] |
Tanaka TU, Rachidi N, Janke C, et al. Evidence that the Ipl1-Sli15(Aurora kinase-INCENP)complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections[J]. Cell, 2002, 108(3):317-329.
doi: 10.1016/S0092-8674(02)00633-5 URL |
| [49] |
Cheeseman IM, Anderson S, Jwa M, et al. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p[J]. Cell, 2002, 111(2):163-172.
pmid: 12408861 |
| [50] |
Gregan J, Polakova S, Zhang LJ, et al. Merotelic kinetochore attachment:causes and effects[J]. Trends Cell Biol, 2011, 21(6):374-381.
doi: 10.1016/j.tcb.2011.01.003 pmid: 21306900 |
| [51] | Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function:centrosomics brings new understanding[J]. Nat Rev Mol Cell Biol, 2007, 8(6):451-463. |
| [52] | Lingle WL, Salisbury JL. The role of the centrosome in the development of malignant tumors[J]. Curr Top Dev Biol, 1999, 49:313-329. |
| [53] |
Brinkley BR. Managing the centrosome numbers game:from chaos to stability in cancer cell division[J]. Trends Cell Biol, 2001, 11(1):18-21.
pmid: 11146294 |
| [54] |
Nigg EA. Centrosome aberrations:cause or consequence of cancer progression?[J]. Nat Rev Cancer, 2002, 2(11):815-825.
doi: 10.1038/nrc924 URL |
| [55] |
Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability[J]. Nature, 2009, 460(7252):278-282.
doi: 10.1038/nature08136 URL |
| [56] |
Wang HF, Takenaka K, Nakanishi A, et al. BRCA2 and nucleophosmin coregulate centrosome amplification and form a complex with the Rho effector kinase ROCK2[J]. Cancer Res, 2011, 71(1):68-77.
doi: 10.1158/0008-5472.CAN-10-0030 URL |
| [57] |
Basto R, Brunk K, Vinadogrova T, et al. Centrosome amplification can initiate tumorigenesis in flies[J]. Cell, 2008, 133(6):1032-1042.
doi: 10.1016/j.cell.2008.05.039 URL |
| [58] |
Habedanck R, Stierhof YD, Wilkinson CJ, et al. The Polo kinase Plk4 functions in centriole duplication[J]. Nat Cell Biol, 2005, 7(11):1140-1146.
pmid: 16244668 |
| [59] |
Meraldi P, Lukas J, Fry AM, et al. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A[J]. Nat Cell Biol, 1999, 1(2):88-93.
pmid: 10559879 |
| [60] |
Wang XX, Yang YL, Duan Q, et al. sSgo1, a major splice variant of Sgo1, functions in centriole cohesion where it is regulated by Plk1[J]. Dev Cell, 2008, 14(3):331-341.
doi: 10.1016/j.devcel.2007.12.007 URL |
| [61] |
Nigg EA. Origins and consequences of centrosome aberrations in human cancers[J]. Int J Cancer, 2006, 119(12):2717-2723.
doi: 10.1002/(ISSN)1097-0215 URL |
| [1] | WANG Zi-ying, LONG Chen-jie, FAN Zhao-yu, ZHANG Lei. Screening of OsCRK5-interacted Proteins in Rice Using Yeast Two-hybrid System [J]. Biotechnology Bulletin, 2023, 39(9): 117-125. |
| [2] | HUANG Xiao-long, SUN Gui-lian, MA Dan-dan, YAN Hui-qing. Construction of Yeast One-hybrid Library and Screening of Factors Regulating LAZY1 Expression in Rice [J]. Biotechnology Bulletin, 2023, 39(9): 126-135. |
| [3] | WEN Xiao-lei, LI Jian-yuan, LI Na, ZHANG Na, YANG Wen-xiang. Construction and Utilization of Yeast Two-hybrid cDNA Library of Wheat Interacted by Puccinia triticina [J]. Biotechnology Bulletin, 2023, 39(9): 136-146. |
| [4] | HAN Hao-zhang, ZHANG Li-hua, LI Su-hua, ZHAO Rong, WANG Fang, WANG Xiao-li. Construction of cDNA Library of Cinnamomun bodinieri Induced by Saline-alkali Stress and Screening of CbP5CS Upstream Regulators [J]. Biotechnology Bulletin, 2023, 39(9): 236-245. |
| [5] | XU Jing, ZHU Hong-lin, LIN Yan-hui, TANG Li-qiong, TANG Qing-jie, WANG Xiao-ning. Cloning of IbHQT1 Promoter and Identification of Upstream Regulatory Factors in Sweet Potato [J]. Biotechnology Bulletin, 2023, 39(8): 213-219. |
| [6] | SONG Zhi-zhong, XU Wei-hua, XIAO Hui-lin, TANG Mei-ling, CHEN Jing-hui, GUAN Xue-qiang, LIU Wan-hao. Cloning, Expression and Function of Iron Regulated Transporter VvIRT1 in Wine Grape(Vitis vinifera L.) [J]. Biotechnology Bulletin, 2023, 39(8): 234-240. |
| [7] | YU Hui, WANG Jing, LIANG Xin-xin, XIN Ya-ping, ZHOU Jun, ZHAO Hui-jun. Isolation and Functional Verification of Genes Responding to Iron and Cadmium Stresses in Lycium barbarum [J]. Biotechnology Bulletin, 2023, 39(7): 195-205. |
| [8] | LI Yu-zhen, MEI Tian-xiu, LI Zhi-wen, WANG Qi, LI Jun, ZOU Yue, ZHAO Xin-qing. Advances in Genomic Studies and Metabolic Engineering of Red Yeasts [J]. Biotechnology Bulletin, 2023, 39(7): 67-79. |
| [9] | HOU Xiao-yuan, CHE Zheng-zheng, LI Heng-jing, DU Chong-yu, XU Qian, WANG Qun-qing. Construction of the Soybean Membrane System cDNA Library and Interaction Proteins Screening for Effector PsAvr3a [J]. Biotechnology Bulletin, 2023, 39(4): 268-276. |
| [10] | WANG Xin-yi, WANG Xiao-qian, WANG Hong-jun, CHAO Yue-hui. Screening, Expression, and Validation of Nanobodies with FLAG Tag [J]. Biotechnology Bulletin, 2023, 39(10): 323-331. |
| [11] | JIANG Nan, SHI Yang, ZHAO Zhi-hui, LI Bin, ZHAO Yi-hui, YANG Jun-biao, YAN Jia-ming, JIN Yu-fan, CHEN Ji, HUANG Jin. Expression and Functional Analysis of OsPT1 Gene in Rice Under Cadmium Stress [J]. Biotechnology Bulletin, 2023, 39(1): 166-174. |
| [12] | CAI Jia, LIANG Zhen-yu, HUANG Yu, LU Yi-shan, SHI gang, JIAN Ji-chang. Screening and Identifing the Interacting Proteins of Grouper(Epinephelus coioides)EcBAG3 Using Yeast Two-hybrid System [J]. Biotechnology Bulletin, 2022, 38(8): 77-83. |
| [13] | YANG Jia-bao, ZHOU Zhi-ming, ZHANG Zhan, FENG Li, SUN Li. Cloning,Expression of Helianthus annuus HaLACS1 Gene and Identification of Its Functional Complementation in Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2022, 38(6): 147-156. |
| [14] | YU Jing, YANG Hui, YU Shi-zhou, ZHAO Hui-na, ZHENG Qing-xia, WANG Bing, LEI Bo. Construction of Yeast One-hybrid Bait Vector of Tobacco NtCBT Gene Promoter and Screening of Interacted Proteins [J]. Biotechnology Bulletin, 2022, 38(10): 73-79. |
| [15] | YE Min, GAO Jiao-qi, ZHOU Yong-jin. Engineering Non-conventional Yeast Cell Factory for the Biosynthesis of Natural Products [J]. Biotechnology Bulletin, 2021, 37(8): 12-24. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||