








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (4): 179-188.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1069
Previous Articles Next Articles
LIU Huan-huan1,2( ), YANG Li-chun1, LI Huo-gen1(
), YANG Li-chun1, LI Huo-gen1( )
)
Received:2023-11-14
Online:2024-04-26
Published:2024-04-30
Contact:
LI Huo-gen
E-mail:lhh91@jsafc.edu.cn;hgli@njfu.edu.cn
LIU Huan-huan, YANG Li-chun, LI Huo-gen. Cloning and Functional Analysis of LtMYB305 in Liriodendron tulipifera[J]. Biotechnology Bulletin, 2024, 40(4): 179-188.
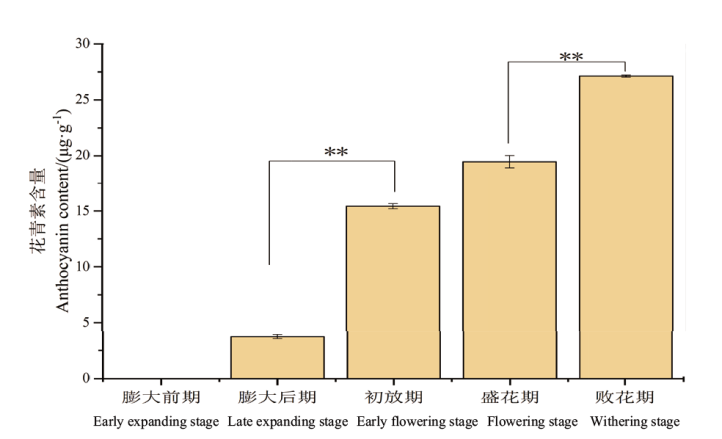
Fig. 1 Anthocyanidin content analysis during floral nectary growth of L. tulipifera * indicates significant difference at P<0.05 level; ** indicates significant difference at P<0.01 level. The same below
| 蛋白 名称 Protein name | 可信度Reliability | |||
|---|---|---|---|---|
| 叶绿体转运肽Chloroplast transit peptide, cTP | 线粒体靶向肽Mitochondrial targeting peptide, mTP | 分泌通路信号肽Secretory pathway signal peptide, SP | 其他 Other | |
| LtMYB305 | 9.40 | 4.90 | 8.00 | 94.70 |
Table 1 Prediction of LtMYB305 protein subcellular localization in L. tulipifera by SignalP-4.0 Server %
| 蛋白 名称 Protein name | 可信度Reliability | |||
|---|---|---|---|---|
| 叶绿体转运肽Chloroplast transit peptide, cTP | 线粒体靶向肽Mitochondrial targeting peptide, mTP | 分泌通路信号肽Secretory pathway signal peptide, SP | 其他 Other | |
| LtMYB305 | 9.40 | 4.90 | 8.00 | 94.70 |
| 蛋白名称 Protein name | 亚细胞结构Subcellular structure | |||
|---|---|---|---|---|
| 细胞核 Nuclear | 过氧化物酶体Peroxisome | 叶绿体Chloroplast | 液泡 Vacuole | |
| MYB305 | 13 | 1 | - | - |
Table 2 Prediction of LtMYB305 protein subcellular localization in L. tulipifera by WOLF PSORT
| 蛋白名称 Protein name | 亚细胞结构Subcellular structure | |||
|---|---|---|---|---|
| 细胞核 Nuclear | 过氧化物酶体Peroxisome | 叶绿体Chloroplast | 液泡 Vacuole | |
| MYB305 | 13 | 1 | - | - |
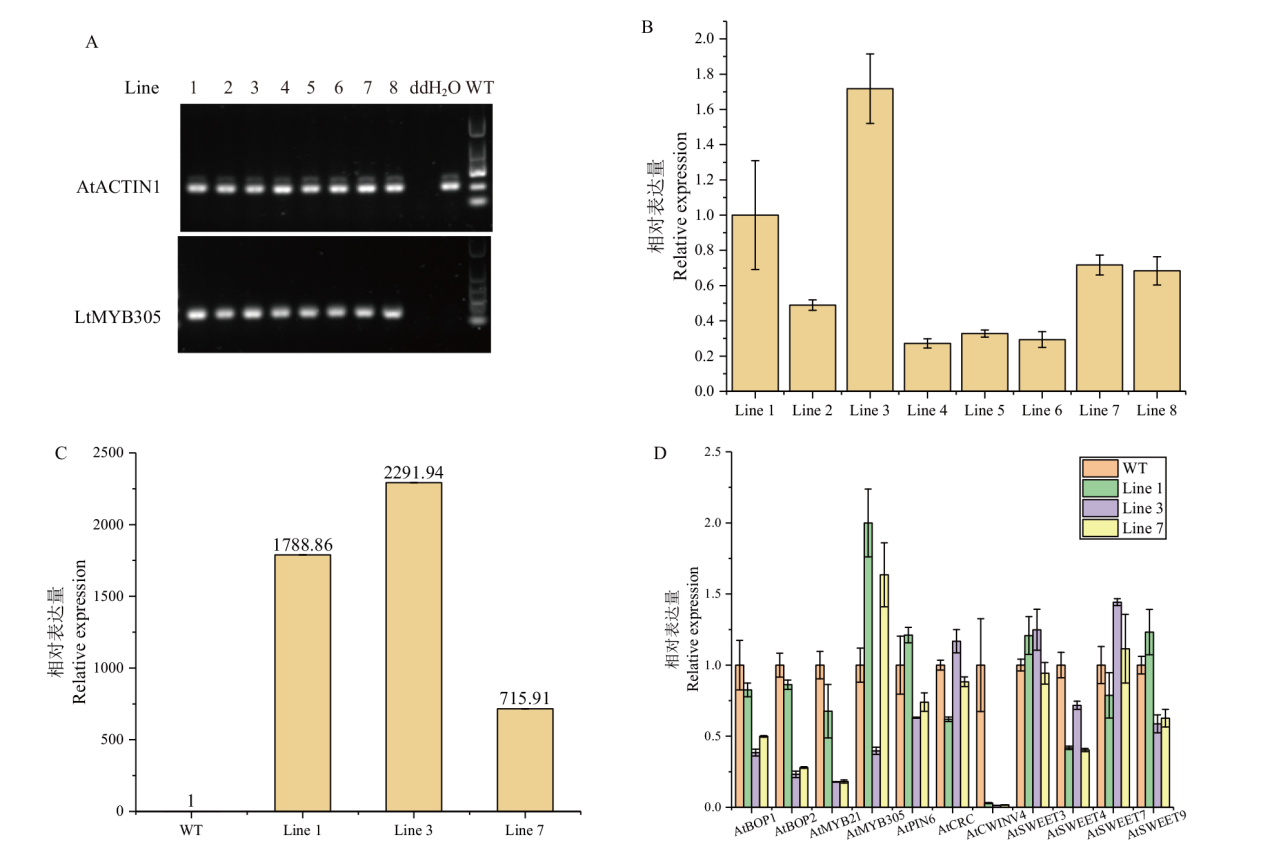
Fig. 5 Expression analysis of T1 generation of positive 35S::LtMYB305-overexpressed A. thaliana plants A: Semi-quantitative PCR analysis in eight lines of T1 generation positive plants; B: RT-qPCR analysis in eight lines of T1 generation positive plants; C: RT-qPCR analysis in three high-expressed lines of T1 generation positive plants; D: RT-qPCR analysis of eleven genes related to nectary development in three high-expressed lines of T1 generation positive plants

Fig. 6 Screening of positive 35S:: LtMYB305-overexpressed A. thaliana plants A: Detection of positive T2-T3 generation 35S::LtMYB305-overexpressed A. thaliana plants. B: Root observation of wild type and 35S::LtMYB305-overexpressed A. thaliana plants. C: Screening of seedling of wild type and 35S::LtMYB305-overexpressed A. thaliana plants

Fig. 7 Phenotype observation of 35S::LtMYB305-overexpressed A. thaliana A: Floral nectaries of wild type A. thaliana(col)plants. B: Floral nectaries 35S::LtMYB305-overexpressed A. thaliana plants. C: T1-T3 generation of 35S::LtMYB305 -overexpressed A. thaliana plants. LN: Lateral nectary
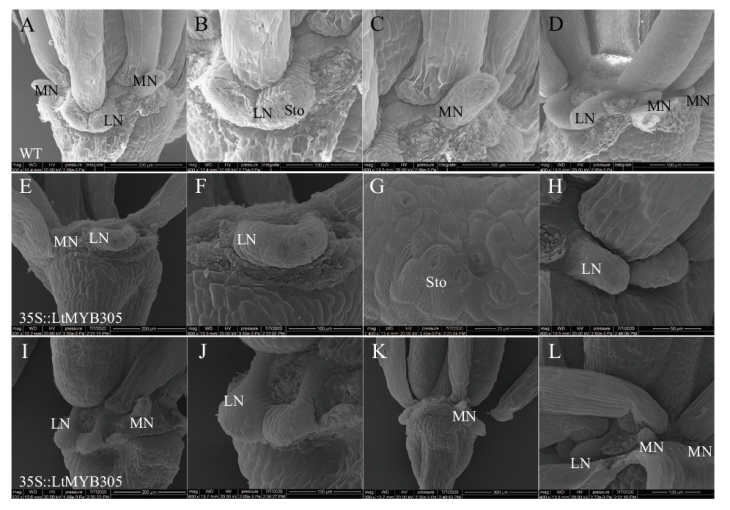
Fig. 9 SEM of floral nectary of 35S::LtMYB305-overexpressed A. thaliana plants A-D: Floral nectaries of wild type A. thaliana(col)plants. E-L: Floral nectaries of 35S::LtMYB305-overexpressed A. thaliana plants. MN: Medial nectary. LN: Lateral nectary. Sto: Stoma
| [1] | Parks CR, Miller NG, Wendel JF, et al. Genetic divergence within the genus Liriodendron(Magnoliaceae)[J]. Ann Mo Bot Gard, 1983, 70(4): 658. |
| [2] |
Hao ZD, Liu SQ, Hu LF, et al. Transcriptome analysis and metabolic profiling reveal the key role of carotenoids in the petal coloration of Liriodendron tulipifera[J]. Hortic Res, 2020, 7: 70.
doi: 10.1038/s41438-020-0287-3 |
| [3] |
Lee JY, Baum SF, Oh SH, et al. Recruitment of CRABS CLAW to promote nectary development within the eudicot clade[J]. Development, 2005, 132(22): 5021-5032.
doi: 10.1242/dev.02067 URL |
| [4] |
Hepworth SR, Zhang YL, McKim S, et al. BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis[J]. Plant Cell, 2005, 17(5): 1434-1448.
doi: 10.1105/tpc.104.030536 pmid: 15805484 |
| [5] |
Nagpal P, Ellis CM, Weber H, et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation[J]. Development, 2005, 132(18): 4107-4118.
doi: 10.1242/dev.01955 pmid: 16107481 |
| [6] |
McKim SM, Stenvik GE, Butenko MA, et al. The BLADE-ON-PETIOLE genes are essential for abscission zone formation in Arabidopsis[J]. Development, 2008, 135(8): 1537-1546.
doi: 10.1242/dev.012807 URL |
| [7] | Reeves PH, Ellis CM, Ploense SE, et al. A regulatory network for coordinated flower maturation[J]. PLoS Genet, 2012, 8(2): e1002506. |
| [8] |
Gross T, Broholm S, Becker A. CRABS CLAW acts as a bifunctional transcription factor in flower development[J]. Front Plant Sci, 2018, 9: 835.
doi: 10.3389/fpls.2018.00835 pmid: 29973943 |
| [9] |
Graf T. MYB: a transcriptional activator linking proliferation and differentiation in hematopoietic cells[J]. Curr Opin Genet Dev, 1992, 2(2): 249-255.
pmid: 1638119 |
| [10] |
Dubos C, Stracke R, Grotewold E, et al. MYB transcription factors in Arabidopsis[J]. Trends Plant Sci, 2010, 15(10): 573-581.
doi: 10.1016/j.tplants.2010.06.005 URL |
| [11] |
Stracke R, Ishihara H, Huep G, et al. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling[J]. Plant J, 2007, 50(4): 660-677.
doi: 10.1111/j.1365-313X.2007.03078.x pmid: 17419845 |
| [12] |
Jackson D, Culianez-Macia F, Prescott AG, et al. Expression patterns of MYB genes from Antirrhinum flowers[J]. Plant Cell, 1991, 3(2): 115-125.
pmid: 1840903 |
| [13] |
Moyano E, Martínez-Garcia JF, Martin C. Apparent redundancy in myb gene function provides gearing for the control of flavonoid biosynthesis in antirrhinum flowers[J]. Plant Cell, 1996, 8(9): 1519-1532.
doi: 10.1105/tpc.8.9.1519 pmid: 8837506 |
| [14] |
Liu GY, Ren G, Guirgis A, et al. The MYB305 transcription factor regulates expression of nectarin genes in the ornamental tobacco floral nectary[J]. Plant Cell, 2009, 21(9): 2672-2687.
doi: 10.1105/tpc.108.060079 URL |
| [15] |
Liu GY, Thornburg RW. Knockdown of MYB305 disrupts nectary starch metabolism and floral nectar production[J]. Plant J, 2012, 70(3): 377-388.
doi: 10.1111/tpj.2012.70.issue-3 URL |
| [16] | Radhika V, Kost C, Boland W, et al. The role of jasmonates in floral nectar secretion[J]. PLoS One, 2010, 5(2): e9265. |
| [17] |
Bender RL, Fekete ML, Klinkenberg PM, et al. PIN6 is required for nectary auxin response and short stamen development[J]. Plant J, 2013, 74(6): 893-904.
doi: 10.1111/tpj.2013.74.issue-6 URL |
| [18] |
Heil M. Induction of two indirect defences benefits Lima bean(Phaseolus Lunatus, Fabaceae)in nature[J]. J Ecol, 2004, 92(3): 527-536.
doi: 10.1111/jec.2004.92.issue-3 URL |
| [19] | Liu HH, Ma JK, Li HG. Transcriptomic and microstructural analyses in Liriodendron tulipifera Linn. reveal candidate genes involved in nectary development and nectar secretion[J]. BMC Plant Biol, 2019, 19(1): 531. |
| [20] | Zhou YW, Li MP, Zhao FF, et al. Floral nectary morphology and proteomic analysis of nectar of Liriodendron tulipifera Linn[J]. Front Plant Sci, 2016, 7: 826. |
| [21] |
Horner HT, Healy RA, Ren G, et al. Amyloplast to chromoplast conversion in developing ornamental tobacco floral nectaries provides sugar for nectar and antioxidants for protection[J]. Am J Bot, 2007, 94(1): 12-24.
doi: 10.3732/ajb.94.1.12 pmid: 21642203 |
| [22] |
Tanaka Y, Sasaki N, Ohmiya A. Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids[J]. Plant J, 2008, 54(4): 733-749.
doi: 10.1111/tpj.2008.54.issue-4 URL |
| [23] |
Wu YQ, Guo J, Zhou Q, et al. De novo transcriptome analysis revealed genes involved in flavonoid biosynthesis, transport and regulation in Ginkgo biloba[J]. Ind Crops Prod, 2018, 124: 226-235.
doi: 10.1016/j.indcrop.2018.07.060 URL |
| [24] |
张月, 袁媛, 何弦, 等. 茉莉花JsMYB108和JsMYB305基因的克隆及其对TPS基因的激活作用[J]. 热带作物学报, 2021, 42(6): 1539-1548.
doi: 10.3969/j.issn.1000-2561.2021.06.005 |
| Zhang Y, Yuan Y, He X, et al. Cloning of JsMYB108 and JsMYB305 and analysis of their activation on TPS gene in Jasmi-num sambac[J]. Chin J Trop Crops, 2021, 42(6): 1539-1548. | |
| [25] |
Davis AR, Pylatuik JD, Paradis JC, et al. Nectar-carbohydrate production and composition vary in relation to nectary anatomy and location within individual flowers of several species of Brassicaceae[J]. Planta, 1998, 205(2): 305-318.
pmid: 9637073 |
| [26] |
Ruhlmann JM, Kram BW, Carter CJ. CELL WALL INVERTASE 4 is required for nectar production in Arabidopsis[J]. J Exp Bot, 2010, 61(2): 395-404.
doi: 10.1093/jxb/erp309 pmid: 19861655 |
| [1] | DU Ze-guang, REN Shao-wen, ZHANG Feng-qin, LI Mei-lan, LI Gai-zhen, QI Xian-hui. Cloning,Expression and Functional Identification of BrMLP328 Gene in Brassica rapa subsp. pekinensis [J]. Biotechnology Bulletin, 2024, 40(4): 122-129. |
| [2] | ZHONG Yun, LIN Chun, LIU Zheng-jie, DONG Chen-wen-hua, MAO Zi-chao, LI Xing-yu. Cloning and Prokaryotic Expression Analysis of Asparagus Saponin Synthesis Related Glycosyltransferase Genes [J]. Biotechnology Bulletin, 2024, 40(4): 255-263. |
| [3] | YANG Yan, HU Yang, LIU Ni-ru, YIN Lu, YANG Rui, WANG Peng-fei, MU Xiao-peng, ZHANG Shuai, CHENG Chun-zhen, ZHANG Jian-cheng. Cloning and Functional Analysis of MbbZIP43 Gene in ‘Hongmantang’ Red-flesh Apple [J]. Biotechnology Bulletin, 2024, 40(2): 146-159. |
| [4] | ZHU Yi, LIU Tang-jing, GONG Guo-yi, ZHANG Jie, WANG Jin-fang, ZHANG Hai-ying. Cloning and Expression Analysis of ClPP2C3 in Citrullus lanatus [J]. Biotechnology Bulletin, 2024, 40(1): 243-249. |
| [5] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [6] | WANG Jia-rui, SUN Pei-yuan, KE Jin, RAN Bin, LI Hong-you. Cloning and Expression Analyses of C-glycosyltransferase Gene FtUGT143 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 204-212. |
| [7] | SUN Ming-hui, WU Qiong, LIU Dan-dan, JIAO Xiao-yu, WANG Wen-jie. Cloning and Expression Analysis of CsTMFs Gene in Tea Plant [J]. Biotechnology Bulletin, 2023, 39(7): 151-159. |
| [8] | LIU Si-jia, WANG Hao-nan, FU Yu-chen, YAN Wen-xin, HU Zeng-hui, LENG Ping-sheng. Cloning and Functional Analysis of LiCMK Gene in Lilium ‘Siberia’ [J]. Biotechnology Bulletin, 2023, 39(3): 196-205. |
| [9] | YANG Xu-yan, ZHAO Shuang, MA Tian-yi, BAI Yu, WANG Yu-shu. Cloning of Three Cabbage WRKY Genes and Their Expressions in Response to Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 261-269. |
| [10] | ZHANG Yu-juan, LI Dong-hua, GONG Hui-hui, CUI Xin-xiao, GAO Chun-hua, ZHANG Xiu-rong, YOU Jun, ZHAO Jun-sheng. Cloning and Salt-tolerance Analysis of NAC Transcription Factor SiNAC77 from Sesamum indicum L. [J]. Biotechnology Bulletin, 2023, 39(11): 308-317. |
| [11] | HOU Rui-ze BAO Yue CHEN Qi-liang MAO Gui-ling WEI Bo-lin HOU Lei-ping LI Mei-lan. Cloning,Expression and Functional Identification of PRR5 Gene in Pakchoi [J]. Biotechnology Bulletin, 2023, 39(10): 128-135. |
| [12] | CHEN Hao-ting, ZHANG Yu-jing, LIU Jie, DAI Ze-min, LIU Wei, SHI Yu, ZHANG Yi, LI Tian-lai. Functional Analysis of WRKY6 Gene in Tomato Under Low-phosphorus Stress [J]. Biotechnology Bulletin, 2023, 39(10): 136-147. |
| [13] | YANG Min, LONG Yu-qing, ZENG Juan, ZENG Mei, ZHOU Xin-ru, WANG Ling, FU Xue-sen, ZHOU Ri-bao, LIU Xiang-dan. Cloning and Function Analysis of Gene UGTPg17 and UGTPg36 in Lonicera macranthoides [J]. Biotechnology Bulletin, 2023, 39(10): 256-267. |
| [14] | LI Xiu-qing, HU Zi-yao, LEI Jian-feng, DAI Pei-hong, LIU Chao, DENG Jia-hui, LIU Min, SUN Ling, LIU Xiao-dong, LI Yue. Cloning and Functional Analysis of Gene GhTIFY9 Related to Cotton Verticillium Wilt Resistance [J]. Biotechnology Bulletin, 2022, 38(8): 127-134. |
| [15] | YU Qiu-lin, MA Jing-yi, ZHAO Pan, SUN Peng-fang, HE Yu-mei, LIU Shi-biao, GUO Hui-hong. Cloning and Functional Analysis of Gynostemma pentaphyllum GpMIR156a and GpMIR166b [J]. Biotechnology Bulletin, 2022, 38(7): 186-193. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||