








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (5): 112-119.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1223
Previous Articles Next Articles
ZHANG Na( ), LIU Meng-nan, QU Zhan-fan, CUI Yi-ping, NI Jia-yao, WANG Hua-zhong(
), LIU Meng-nan, QU Zhan-fan, CUI Yi-ping, NI Jia-yao, WANG Hua-zhong( )
)
Received:2023-12-29
Online:2024-05-26
Published:2024-03-21
Contact:
WANG Hua-zhong
E-mail:zn1999227@126.com;skywhz@tjnu.edu.cn
ZHANG Na, LIU Meng-nan, QU Zhan-fan, CUI Yi-ping, NI Jia-yao, WANG Hua-zhong. Alternative Translation of Wheat Enolase-encoding Gene ENO2 and Its Prokaryotic Expression[J]. Biotechnology Bulletin, 2024, 40(5): 112-119.
| 引物名称 Prime name | 引物序列 Primer sequence(5'-3') |
|---|---|
| ENO-m-F | AAGTCCGGAGCTAGCTATGGCGGCGACGATCCAGTC |
| ENO-m-R | GCCCTTGCTCACCATGGCGTATGGCTCCACCGGTGCAC |
| Δ93-m-F | AAGTCCGGAGCTAGCTATGGTTCAGCAGCTTGATGGA |
| Δ93-m-R | GCCCTTGCTCACCATGGCGTATGGCTCCACCGGTGCAC |
| M94A-F1 | AAGTCCGGAGCTAGCTATGGCGGCGACGATCCAGTC |
| M94A-R1 | AACAGCAAAGTTGTCGAGCTCAGTTTGAG |
| M94A-F2 | GCTCGACAACTTTGCTGTTCAGCAGCTTGATGGAACCAAG |
| M94A-R2 | GCCCTTGCTCACCATGGCGTATGGCTCCACCGGTGCAC |
| ENO-GST-F | GATCTGGTTCCGCGTGGATCCATGGCGGCGACGATCCAGTC |
| ENO-GST-R | GTCACGATGCGGCCGCTCGAGGTATGGCTCCACCGGTGCAC |
Table 1 Primers used in this study
| 引物名称 Prime name | 引物序列 Primer sequence(5'-3') |
|---|---|
| ENO-m-F | AAGTCCGGAGCTAGCTATGGCGGCGACGATCCAGTC |
| ENO-m-R | GCCCTTGCTCACCATGGCGTATGGCTCCACCGGTGCAC |
| Δ93-m-F | AAGTCCGGAGCTAGCTATGGTTCAGCAGCTTGATGGA |
| Δ93-m-R | GCCCTTGCTCACCATGGCGTATGGCTCCACCGGTGCAC |
| M94A-F1 | AAGTCCGGAGCTAGCTATGGCGGCGACGATCCAGTC |
| M94A-R1 | AACAGCAAAGTTGTCGAGCTCAGTTTGAG |
| M94A-F2 | GCTCGACAACTTTGCTGTTCAGCAGCTTGATGGAACCAAG |
| M94A-R2 | GCCCTTGCTCACCATGGCGTATGGCTCCACCGGTGCAC |
| ENO-GST-F | GATCTGGTTCCGCGTGGATCCATGGCGGCGACGATCCAGTC |
| ENO-GST-R | GTCACGATGCGGCCGCTCGAGGTATGGCTCCACCGGTGCAC |

Fig. 1 Protein sequence alignment of wheat and Arabidopsis enolases TaENO2-5A, TaENO2-5B, and TaENO2-5D are wheat enolases of homeologous ENO2 proteins, and AtENO1, AtENO2, and AtENO3 are Arabidopsis enolase proteins. Solid red box indicates the serine residue(S)responsible for binding of Mg2+ in the catalytic center. Dashed red box indicates the second methionine residue(M)in the ENO2 sequences. The ATG codon encoding this residue is an internal start codon of alternative translation

Fig. 2 Alternative translation products of TaENO2 A: Schematic representation of the three forms(I, II, and III)of mCherry-fused TaENO2 coding sequences. B: Subcellular localization of the alternative translation products of TaENO2. Different mCherry fusion genes shown in(A)were respectively transformed into protoplasts for expression. The subcellular distribution of red fluorescence and the Hoechst 33342-stained nuclei in protoplasts were observed at 24 h after protoplast transformation. Scale bar indicates 10 μm. C: Western blot analysis on the alternative translation products of TaENO2 expressed in protoplasts
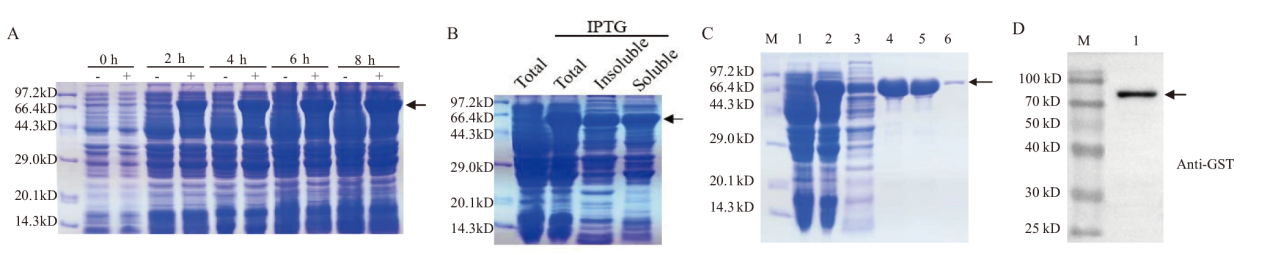
Fig. 3 Prokaryotic expression and purification of recombinant GST-TaENO2 protein A: Prokaryotic expression of recombinant GST-TaENO2 protein. The E. coli strain BL21(DE3)carrying the fusion gene GST-TaENO2 was grown to the log growth phase. The heterologous protein expression was then induced with IPTG for the indicated time periods. Cells were lysed by boiling and an equal aliquot of protein was resolved by SDS-PAGE. Arrows indicate the bands of recombinant protein. B: Solubility analysis on the prokaryotically expressed recombinant GST-TaENO2 protein. E. coli culture induced by IPTG was lysed by sonication. The soluble and insoluble protein fractions were separated by centrifugation and resolved by SDS-PAGE. Arrows indicate the bands of recombinant protein GST-TaENO2. C: Purification of recombinant GST-TaENO2 protein. M: Protein marker; 1: Total extracted protein from the E. coli culture which was not subjected to IPTG induction of gene expression; 2: Total extracted protein from the E.coli culture which had been subjected to IPTG induction of gene expression for 4 h; 3-6: Collected flow-through(3)and successive elution fractions(4-6)in the process of protein purification by affinity chromatography. D: Western blot analysis of the purified recombinant GST-TaENO2 protein. Arrows indicate the bands of recombinant protein
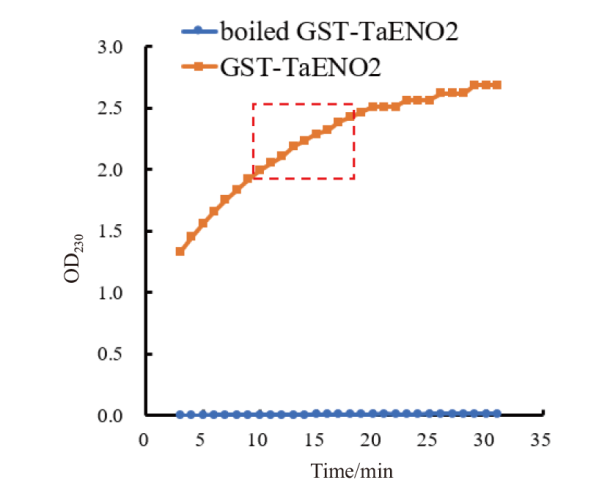
Fig. 4 In vitro assays on the enolase activity of the recombinant protein GST-TaENO2 The catalytic product of enolase, PEP, has maximum optical absorption at a wavelength of 230 nm. The figure shows the time-dependent changes in the optical density of the in vitro enolase assay solutions at 230 nm(OD230). Red dash box indicates the near linear increase stage of OD230. The data of this stage were used for calculation of enolase activity
| [1] |
Andriotis VME, Kruger NJ, et al. Plastidial glycolysis in developing Arabidopsis embryos[J]. New Phytol, 2010, 185(3): 649-662.
doi: 10.1111/j.1469-8137.2009.03113.x pmid: 20002588 |
| [2] |
Lee H, Guo Y, Ohta M, et al. LOS2, a genetic locus required for cold-responsive gene transcription encodes a bi-functional enolase[J]. EMBO J, 2002, 21(11): 2692-2702.
doi: 10.1093/emboj/21.11.2692 pmid: 12032082 |
| [3] |
Voll LM, Hajirezaei MR, Czogalla-Peter C, et al. Antisense inhibition of enolase strongly limits the metabolism of aromatic amino acids, but has only minor effects on respiration in leaves of transgenic tobacco plants[J]. New Phytol, 2009, 184(3): 607-618.
doi: 10.1111/j.1469-8137.2009.02998.x pmid: 19694966 |
| [4] |
Troncoso-Ponce MA, Rivoal J, Dorion S, et al. Molecular and biochemical characterization of the sunflower(Helianthus annuus L.) cytosolic and plastidial enolases in relation to seed development[J]. Plant Sci, 2018, 272: 117-130.
doi: S0168-9452(18)30173-0 pmid: 29807582 |
| [5] |
Qiao G, Wu AG, Chen XL, et al. Enolase 1, a moonlighting protein, as a potential target for cancer treatment[J]. Int J Biol Sci, 2021, 17(14): 3981-3992.
doi: 10.7150/ijbs.63556 pmid: 34671213 |
| [6] | Liu WC, Song RF, Qiu YM, et al. Sulfenylation of ENOLASE2 facilitates H2O2-conferred freezing tolerance in Arabidopsis[J]. Dev Cell, 2022, 57(15): 1883-1898.e5. |
| [7] | Barkla BJ, Vera-Estrella R, et al. Quantitative proteomics of the tonoplast reveals a role for glycolytic enzymes in salt tolerance[J]. Plant Cell, 2009, 21(12): 4044-4058. |
| [8] | Liu ZJ, Zheng LM, Pu L, et al. ENO2 affects the seed size and weight by adjusting cytokinin content and forming ENO2-bZIP75 complex in Arabidopsis thaliana[J]. Front Plant Sci, 2020, 11: 574316. |
| [9] | Liu ZJ, Liu HM, Zheng LM, et al. Enolase2 regulates seed fatty acid accumulation via mediating carbon partitioning in Arabidopsis thaliana[J]. Physiol Plant, 2022, 174(6): e13797. |
| [10] |
Zhang YJ, Sampathkumar A, Kerber SML, et al. A moonlighting role for enzymes of glycolysis in the co-localization of mitochondria and chloroplasts[J]. Nat Commun, 2020, 11(1): 4509.
doi: 10.1038/s41467-020-18234-w pmid: 32908151 |
| [11] | Eremina M, Rozhon W, et al. ENO2 activity is required for the development and reproductive success of plants, and is feedback-repressed by AtMBP-1[J]. Plant J, 2015, 81(6): 895-906. |
| [12] | Liu ZJ, Zhang YH, Ma XF, et al. Biological functions of Arabidopsis thaliana MBP-1-like protein encoded by ENO2 in the response to drought and salt stresses[J]. Physiol Plant, 2020, 168(3): 660-674. |
| [13] | Yang LY, Wang ZX, Zhang AQ, et al. Reduction of the canonical function of a glycolytic enzyme enolase triggers immune responses that further affect metabolism and growth in Arabidopsis[J]. Plant Cell, 2022, 34(5): 1745-1767. |
| [14] | Ma XF, Wu Y, Ming HN, et al. AtENO2 functions in the development of male gametophytes in Arabidopsis thaliana[J]. J Plant Physiol, 2021, 263: 153417. |
| [15] | Bi CX, Yu YH, Dong CH, et al. The bZIP transcription factor TabZIP15 improves salt stress tolerance in wheat[J]. Plant Biotechnol J, 2021, 19(2): 209-211. |
| [16] | Zhang YF, Wang JY, et al. Wheat TaSnRK2.10 phosphorylates TaERD15 and TaENO1 and confers drought tolerance when overexpressed in rice[J]. Plant Physiol, 2023, 191(2): 1344-1364. |
| [17] | Liu ZJ, Zhang A, Zheng LM, et al. The biological significance and regulatory mechanism of c-myc binding protein 1(MBP-1)[J]. Int J Mol Sci, 2018, 19(12): 3868. |
| [18] | Kang M, Abdelmageed H, Lee S, et al. AtMBP-1, an alternative translation product of LOS2, affects abscisic acid responses and is modulated by the E3 ubiquitin ligase AtSAP5[J]. Plant J, 2013, 76(3): 481-493. |
| [19] |
杜晓敏, 王均, 等. 小麦叶肉细胞原生质体制备参数解析及在基因瞬时表达上的应用[J]. 华北农学报, 2015, 30(6): 52-60.
doi: 10.7668/hbnxb.2015.06.008 |
| Du XM, Wang J, et al. Factors influencing the preparation of wheat mesophyll protoplasts and the application of wheat mesophyll protoplasts in gene transient expression[J]. Acta Agric Boreali Sin, 2015, 30(6): 52-60. |
| [1] | PAN Ping-ping, XU Zhi-hao, ZHANG Yi-wen, LI Qing, WANG Zhong-hua. Prokaryotic Expression, Subcellular Localization and Expression Analysis of PcCHS Gene from Polygonatum cyrtonema Hua [J]. Biotechnology Bulletin, 2024, 40(5): 280-289. |
| [2] | ZHONG Yun, LIN Chun, LIU Zheng-jie, DONG Chen-wen-hua, MAO Zi-chao, LI Xing-yu. Cloning and Prokaryotic Expression Analysis of Asparagus Saponin Synthesis Related Glycosyltransferase Genes [J]. Biotechnology Bulletin, 2024, 40(4): 255-263. |
| [3] | JIAO Jin-lan, WANG Wen-wen, JIE Xin-rui, WANG Hua-zhong, YUE Jie-yu. Mechanism of Exogenous Calcium Alleviating Salt Stress Toxicity in Wheat Seedlings [J]. Biotechnology Bulletin, 2024, 40(1): 207-221. |
| [4] | CHANG Lu-yin, WANG Zhong-hua, LI Feng-min, GAO Zi-yuan, ZHANG Hui-hong, WANG Yi, LI Fang, HAN Yan-lai, JIANG Ying. Screening Multi-functional Rhizobacteria from Maize Rhizosphere and Their Ehancing Effects on Winter Wheat-Summer Maize Rotation System [J]. Biotechnology Bulletin, 2024, 40(1): 231-242. |
| [5] | WEN Xiao-lei, LI Jian-yuan, LI Na, ZHANG Na, YANG Wen-xiang. Construction and Utilization of Yeast Two-hybrid cDNA Library of Wheat Interacted by Puccinia triticina [J]. Biotechnology Bulletin, 2023, 39(9): 136-146. |
| [6] | HAN Zhi-yang, JIA Zi-miao, LIANG Qiu-ju, WANG Ke, TANG Hua-li, YE Xing-guo, ZHANG Shuang-xi. Salt Tolerance at Seedling Stage and Analysis of Selenium and Folic Acid Content in Seeds in Two Sets of Wheat-Dasypyrum villosum Chromosom Additional Lines [J]. Biotechnology Bulletin, 2023, 39(8): 185-193. |
| [7] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [8] | WANG Jia-rui, SUN Pei-yuan, KE Jin, RAN Bin, LI Hong-you. Cloning and Expression Analyses of C-glycosyltransferase Gene FtUGT143 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 204-212. |
| [9] | MEI Huan, LI Yue, LIU Ke-meng, LIU Ji-hua. Study on the Biosynthesis of l-SLR by Efficient Prokaryotic Expression of Berberine Bridge Enzyme [J]. Biotechnology Bulletin, 2023, 39(7): 277-287. |
| [10] | LIU Hui, LU Yang, YE Xi-miao, ZHOU Shuai, LI Jun, TANG Jian-bo, CHEN En-fa. Comparative Transcriptome Analysis of Cadmium Stress Response Induced by Exogenous Sulfur in Tartary Buckwheat [J]. Biotechnology Bulletin, 2023, 39(5): 177-191. |
| [11] | TENG Meng-xin, XU Ya, HE Jing, WANG Qi, QIAO Fei, LI Jing-yang, LI Xin-guo. Cloning and Prokaryotic Expression Analysis of MaMC6 in Banana [J]. Biotechnology Bulletin, 2023, 39(12): 179-186. |
| [12] | KONG De-zhen, NIE Ying-bin, CUI Feng-juan, SANG Wei, XU Hong-jun, TIAN Xiao-ming. Research Status and Prospect of Hybrid Wheat Seed Production [J]. Biotechnology Bulletin, 2023, 39(1): 95-103. |
| [13] | SUO Qing-qing, WU Nan, YANG Hui, LI Li, WANG Xi-feng. Prokaryotic Expression,Antibody Preparation and Application of Rice Caffeoyl Coenzyme A-O-methyltransferase Gene [J]. Biotechnology Bulletin, 2022, 38(8): 135-141. |
| [14] | QIN Xue-jing, WANG Yu-han, CAO Yi-bo, ZHANG Ling-yun. Prokaryotic Expression and Preparation of Polyclonal Antibody of PwHAP5 Gene in Picea wilsonii [J]. Biotechnology Bulletin, 2022, 38(8): 142-149. |
| [15] | WANG Guang-li, FAN Chan, WANG Hui, LU Hui-fang, XIA Ling-yin, HUANG Jian, MIN Xun. Prokaryotic Expression,Purification,Identification,and Polyclonal Antibody Preparation of Vibrio cholerae Hemolysin HlyA [J]. Biotechnology Bulletin, 2022, 38(7): 269-277. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||