








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (7): 197-206.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0039
Previous Articles Next Articles
LIAO Yang-mei1( ), ZHAO Guo-chun1, WENG Xue-huang2, JIA Li-ming1, CHEN Zhong1(
), ZHAO Guo-chun1, WENG Xue-huang2, JIA Li-ming1, CHEN Zhong1( )
)
Received:2024-01-12
Online:2024-07-26
Published:2024-05-24
Contact:
CHEN Zhong
E-mail:2414729525@qq.com;zhongchen@bjfu.edu.cn
LIAO Yang-mei, ZHAO Guo-chun, WENG Xue-huang, JIA Li-ming, CHEN Zhong. Transcriptome Sequencing of Male Sterile Buds at Different Developmental Stages in Sapindus mukorossi ‘Qirui’[J]. Biotechnology Bulletin, 2024, 40(7): 197-206.
| 基因编号Gene ID | 基因名称Gene name | 正向引物序列Forward primer sequence(5'-3') | 反向引物序列Reverse primer sequence(5'-3') |
|---|---|---|---|
| Samuk01G0221000 | SmDYT1 | CACTTACATAGAGGTGCTG | CTCAATCCCACTATTCTTC |
| Samuk09G0006500 | SmMYB35 | ACATACCTCACCACCACCT | TTGACCATCTTCTGCCTAT |
| Samuk03G0239600 | SmbHLH091 | ACGGCAGGAAGATGTCAGT | GTCGTGAATCAAATGGGTG |
| Samuk02G0196100 | SmCAT2 | AACCCAAAGTCTCACATCC | CACCCACATCATCAAAGAG |
| Samuk12G0082500 | SmMIOX1 | TGCTGAGGCTATTAGGAAG | CGAAGTTAGGCAGAAGAAG |
| Samuk07G0073400 | SmCYP704B1 | AAAAGCAGGAGGGATGGTG | TGTATGAGGCTGCGTCAGG |
| Samuk07G0100300 | SmMYB62 | AACAGCCAAATCACAGACG | GCTCCTAGATTGAACCCAC |
| Samuk03G0033700 | SmTCF1 | TTAAGTCCAATATGCGTGTC | GCCCAAACTGATTCCCAC |
| Samuk04G0015800 | SmCER1 | GGCATCCAGGTTACCACAT | CATCTCCCACCACCCATAC |
| Samuk07G0011500 | SmRPL1 | CTGACCACCCCACCACTCTC | TCCTCCTCATCTGCCACCTC |
Table 1 Primers used in the RT-qPCR
| 基因编号Gene ID | 基因名称Gene name | 正向引物序列Forward primer sequence(5'-3') | 反向引物序列Reverse primer sequence(5'-3') |
|---|---|---|---|
| Samuk01G0221000 | SmDYT1 | CACTTACATAGAGGTGCTG | CTCAATCCCACTATTCTTC |
| Samuk09G0006500 | SmMYB35 | ACATACCTCACCACCACCT | TTGACCATCTTCTGCCTAT |
| Samuk03G0239600 | SmbHLH091 | ACGGCAGGAAGATGTCAGT | GTCGTGAATCAAATGGGTG |
| Samuk02G0196100 | SmCAT2 | AACCCAAAGTCTCACATCC | CACCCACATCATCAAAGAG |
| Samuk12G0082500 | SmMIOX1 | TGCTGAGGCTATTAGGAAG | CGAAGTTAGGCAGAAGAAG |
| Samuk07G0073400 | SmCYP704B1 | AAAAGCAGGAGGGATGGTG | TGTATGAGGCTGCGTCAGG |
| Samuk07G0100300 | SmMYB62 | AACAGCCAAATCACAGACG | GCTCCTAGATTGAACCCAC |
| Samuk03G0033700 | SmTCF1 | TTAAGTCCAATATGCGTGTC | GCCCAAACTGATTCCCAC |
| Samuk04G0015800 | SmCER1 | GGCATCCAGGTTACCACAT | CATCTCCCACCACCCATAC |
| Samuk07G0011500 | SmRPL1 | CTGACCACCCCACCACTCTC | TCCTCCTCATCTGCCACCTC |

Fig. 1 Microstructure of early development of ‘Qirui’ anther T1, T2 and T3 refer to the 8th(S8), 9th(S9)and 10b(S10b)stages of S. mukorossi Gaertn. flower development respectively, and S12 refers to the 12th stage. T: Tapetum; Tds: tetrad; E: epidermis; En: endothecium; ML: middle layer. Bar = 100 μm. The same below
| 样本名称 Sample name | 原始数据 Raw reads | 过滤后数据 Total clean reads | 总映射率 Total mapping ratio/% | 单一映射率 Uniq mapped reads/% | Q20/% | Q30/% |
|---|---|---|---|---|---|---|
| T1-1 | 88 252 858 | 85 911 910 | 95.86 | 72.67 | 98.37 | 95.02 |
| T1-2 | 91 157 116 | 88 898 424 | 95.83 | 82.08 | 98.46 | 95.22 |
| T1-3 | 88 984 514 | 86 405 806 | 95.85 | 75.65 | 98.10 | 94.31 |
| T2-1 | 88 937 894 | 86 784 106 | 95.91 | 80.89 | 98.28 | 94.76 |
| T2-2 | 100 745 016 | 98 376 588 | 95.87 | 82.46 | 98.42 | 95.11 |
| T2-3 | 107 473 734 | 105 653 102 | 95.97 | 84.99 | 98.44 | 95.12 |
| T3-1 | 95 388 566 | 93 391 938 | 95.77 | 86.11 | 98.36 | 94.95 |
| T3-2 | 89 881 480 | 87 909 282 | 96.08 | 80.18 | 98.43 | 95.17 |
| T3-3 | 96 582 048 | 95 072 204 | 96.00 | 84.08 | 98.39 | 95.00 |
Table 2 Data evaluation of sample sequencing
| 样本名称 Sample name | 原始数据 Raw reads | 过滤后数据 Total clean reads | 总映射率 Total mapping ratio/% | 单一映射率 Uniq mapped reads/% | Q20/% | Q30/% |
|---|---|---|---|---|---|---|
| T1-1 | 88 252 858 | 85 911 910 | 95.86 | 72.67 | 98.37 | 95.02 |
| T1-2 | 91 157 116 | 88 898 424 | 95.83 | 82.08 | 98.46 | 95.22 |
| T1-3 | 88 984 514 | 86 405 806 | 95.85 | 75.65 | 98.10 | 94.31 |
| T2-1 | 88 937 894 | 86 784 106 | 95.91 | 80.89 | 98.28 | 94.76 |
| T2-2 | 100 745 016 | 98 376 588 | 95.87 | 82.46 | 98.42 | 95.11 |
| T2-3 | 107 473 734 | 105 653 102 | 95.97 | 84.99 | 98.44 | 95.12 |
| T3-1 | 95 388 566 | 93 391 938 | 95.77 | 86.11 | 98.36 | 94.95 |
| T3-2 | 89 881 480 | 87 909 282 | 96.08 | 80.18 | 98.43 | 95.17 |
| T3-3 | 96 582 048 | 95 072 204 | 96.00 | 84.08 | 98.39 | 95.00 |
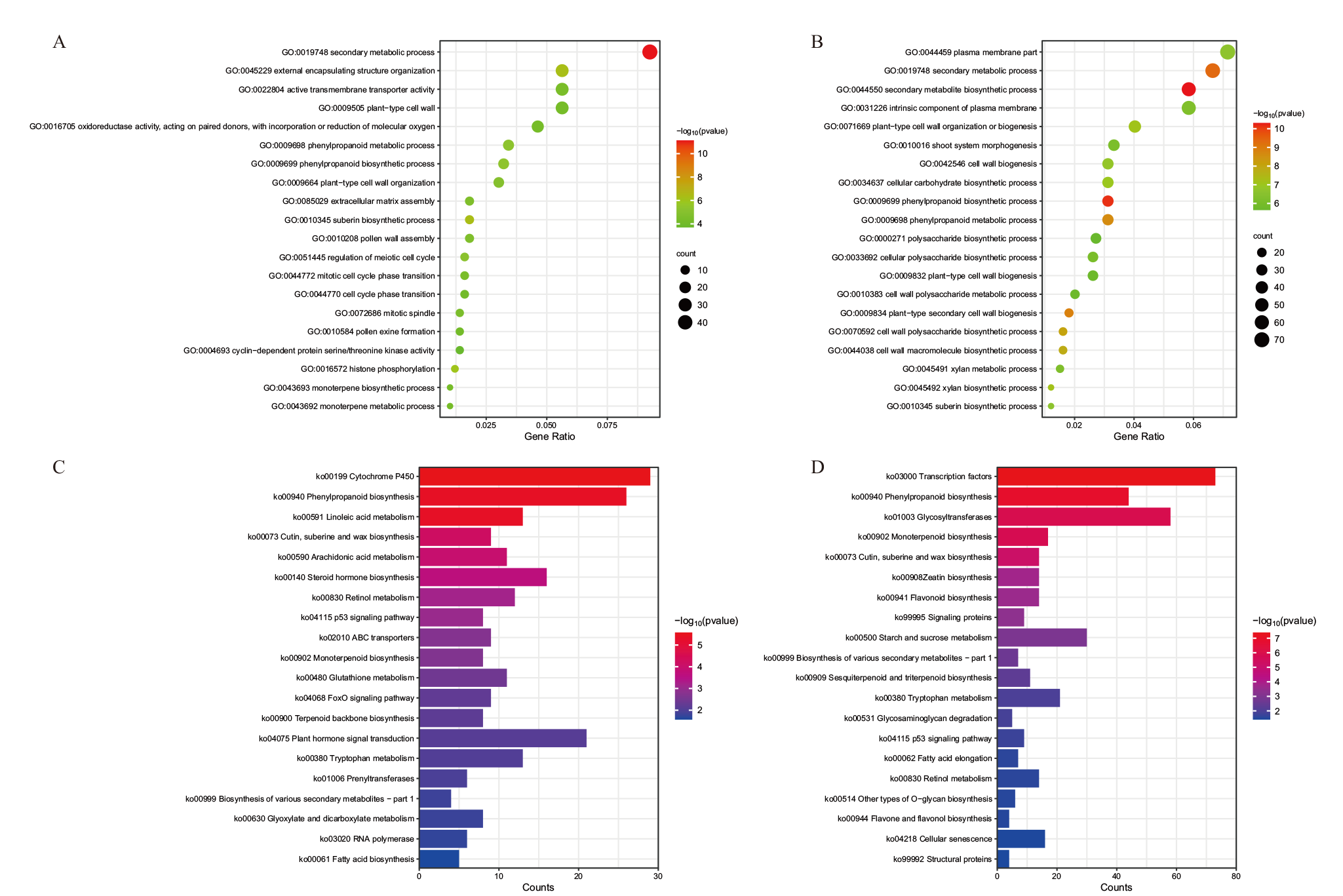
Fig. 5 GO functional enrichment and KEGG pathway enrichment of differentially expressed genes A, C: GO functional annotation(A)and KEGG pathway enrichment(C)of differentially expressed genes in T2_vs_T1; B, D: GO functional annotation(B)and KEGG pathway enrichment(D)of differentially expressed genes in T3_vs_T2
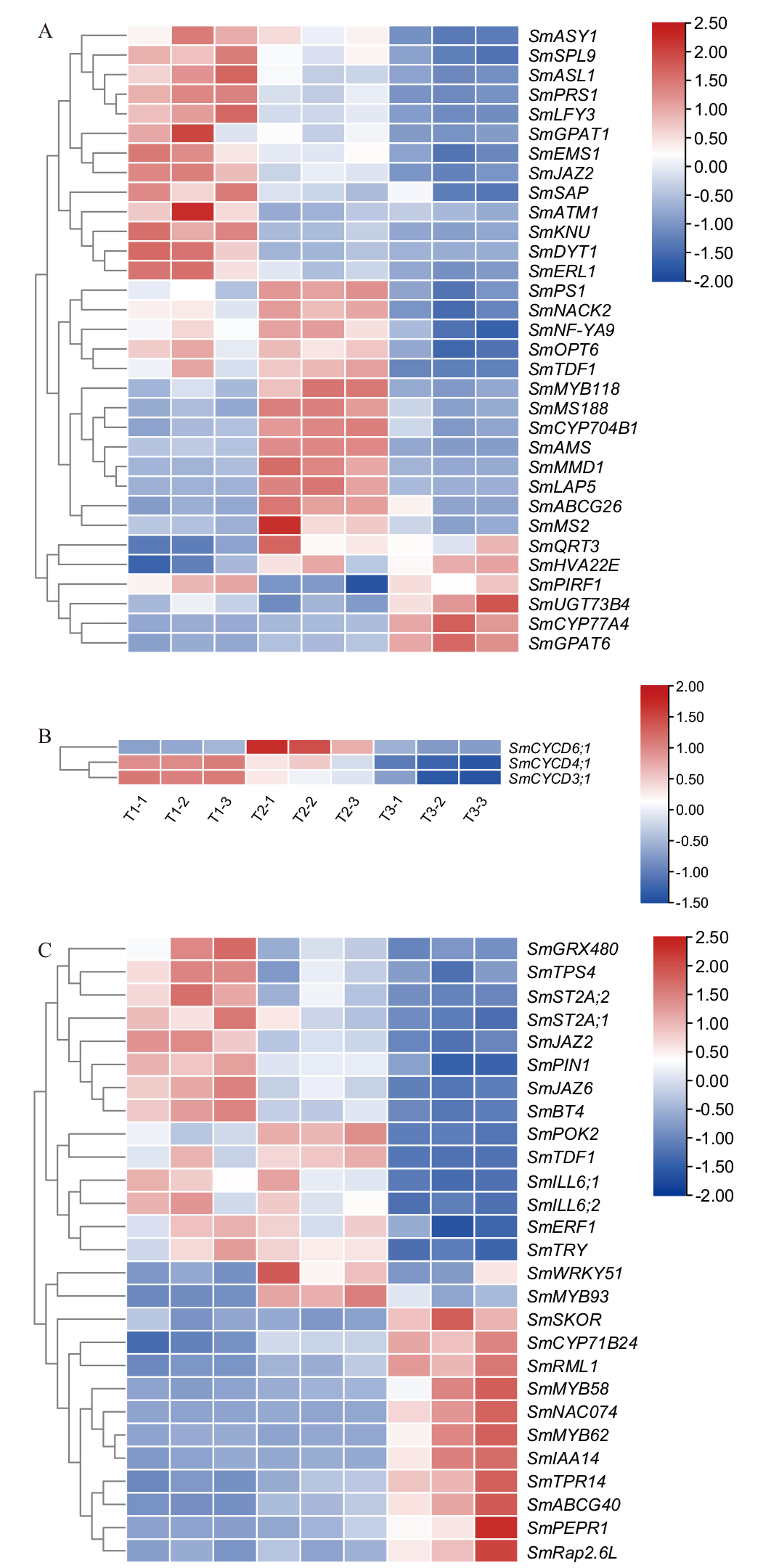
Fig. 6 Expression analysis of key candidate differential genes for male sterility A: Pollen development genes. B: Cell division CYCD family of 3 genes. C: Jasmonic acid synthesis and signal transduction genes
| [1] | Xu YS, Yu D, Chen J, et al. A review of rice male sterility types and their sterility mechanisms[J]. Heliyon, 2023. |
| [2] | 李翔, 李涛, 宫超, 等. 番茄雄性不育研究现状与展望[J]. 广东农业科学, 2023, 50(02): 49-58. |
| Li X, Li T, Gong C, et al. Current situation and prospect of research on male sterility of tomato[J]. Guangdong Agricultural Sciences, 2023, 50(02): 49-58. | |
| [3] | Scott RJ, Spielman M, Dickinson HG. Stamen structure and function[J]. The Plant Cell, 2004, 16(suppl_1): S46-S60. |
| [4] | Kaul MLH. Male sterility in higher plants[M]. Springer Science & Business Media, 2012. |
| [5] |
Chen L, Liu YG. Male sterility and fertility restoration in crops[J]. Annual review of plant biology, 2014, 65: 579-606.
doi: 10.1146/annurev-arplant-050213-040119 pmid: 24313845 |
| [6] | De Storme N, Geelen D. The impact of environmental stress on male reproductive development in plants: biological processes and molecular mechanisms[J]. Plant Cell Environ, 2014, 37(1): 1-18. |
| [7] |
Dukowic-Schulze S, van der Linde K. Oxygen, secreted proteins and small RNAs: mobile elements that govern anther development[J]. Plant Reproduction, 2021, 34(1): 1-19.
doi: 10.1007/s00497-020-00401-0 pmid: 33492519 |
| [8] | 吴智明, 胡开林, 符积钦, 等. 辣椒胞质雄性不育与花蕾内源激素含量的关系[J]. 华南农业大学学报, 2010, 31(2): 1-4. |
| Wu ZM, Hu KL, Fu JQ, et al. Relationships between cytoplasmic male sterility and endogenous hormone content of pepper bud[J]. Journal of South China Agricultural University, 2010, 31(2): 1-4. | |
| [9] | Liu H, Xie WF, Zhang L, et al. Auxin biosynthesis by the YUCCA6 flavin monooxygenase gene in woodland strawberry[J]. Journal of Integrative Plant Biology, 2014, 56(4): 350-363. |
| [10] | Zhang ZB, Zhu J, Gao JF, et al. Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis[J]. The Plant Journal, 2007, 52(3): 528-538. |
| [11] | Thorstensen T, Grini PE, Mercy IS, et al. The Arabidopsis SET-domain protein ASHR3 is involved in stamen development and interacts with the bHLH transcription factor ABORTED MICROSPORES(AMS)[J]. Plant Molecular Biology, 2008, 66(1): 47-59. |
| [12] | Xiong SX, Lu JY, Lou Y, et al. The transcription factors MS 188 and AMS form a complex to activate the expression of CYP703A2 for sporopollenin biosynthesis in Arabidopsis thaliana[J]. The Plant Journal, 2016, 88(6): 936-946. |
| [13] | 刘济铭, 孙操稳, 何秋阳, 等. 国内外无患子属种质资源研究进展[J]. 世界林业研究, 2017, 30(6): 12-18. |
| Liu JM, Sun CW, He QY, et al. Research progress in Sapindus L. germplasm resources[J]. World Forestry Research, 2017, 30(6): 12-18. | |
| [14] | 邵文豪, 刁松锋, 董汝湘, 等. 无患子种实形态及经济性状的地理变异[J]. 林业科学研究, 2013, 26(5): 603-608. |
| Shao WH, Diao SF, Dong RX, et al. Study on geographic variation of morphology and economic character of fruit and seed of Sapindus mukorossi[J]. Forest Research, 2013, 26(5): 603-608. | |
| [15] | 赵国春. 无患子花发育及性别分化机理研究[D]. 北京: 北京林业大学, 2023. |
| Zhao GC. Study on the mechanism of flower development and sex differentiation of Sapindus mukorossi[D]. Beijing: Beijing Forestry University, 2023. | |
| [16] | Xu YY, Zhao GC, Ji XQ, et al. Metabolome and transcriptome analysis reveals the transcriptional regulatory mechanism of triterpenoid saponin biosynthesis in soapberry(Sapindus mukorossi Gaertn.)[J]. Journal of Agricultural and Food Chemistry, 2022, 70(23): 7095-7109. |
| [17] |
徐圆圆, 赵国春, 郝颖颖, 等. 无患子RT-qPCR内参基因的筛选与验证[J]. 生物技术通报, 2022, 38(10): 80-89.
doi: 10.13560/j.cnki.biotech.bull.1985.2021-1616 |
| Xu YY, Zhao GC, Hao YY, et al. Reference genes selection and validation for RT-qPCR in Sapindus mukorossi[J]. Biotechnology Bulletin, 2022, 38(10): 80-89. | |
| [18] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCT method[J]. Methods, 2001, 25(4): 402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [19] |
Jiang J, Zhang Z, Cao J. Pollen wall development: the associated enzymes and metabolic pathways[J]. Plant Biology, 2013, 15(2): 249-263.
doi: 10.1111/j.1438-8677.2012.00706.x pmid: 23252839 |
| [20] | Vu KV, Nguyen NT, Jeong CY, et al. Systematic deletion of the ER lectin chaperone genes reveals their roles in vegetative growth and male gametophyte development in Arabidopsis[J]. The Plant Journal, 2017, 89(5): 972-983. |
| [21] |
Yang K, Zhou XJ, Wang YY, et al. Carbohydrate metabolism and gene regulation during anther development in an androdioecious tree, Tapiscia sinensis[J]. Annals of Botany, 2017, 120(6): 967-977.
doi: 10.1093/aob/mcx094 pmid: 28961748 |
| [22] | Wang R, Dobritsa AA. Exine and aperture patterns on the pollen surface: their formation and roles in plant reproduction[J]. Annual Plant Reviews Online, 2018: 589-628. |
| [23] | Mondol PC, Xu D, Duan L, et al. Defective pollen wall 3(DPW3), a novel alpha integrin-like protein, is required for pollen wall formation in rice[J]. New Phytologist, 2020, 225(2): 807-822. |
| [24] | Li FS, Phyo P, Jacobowitz J, et al. The molecular structure of plant sporopollenin[J]. Nature plants, 2019, 5(1): 41-46. |
| [25] | Tidy AC, Ferjentsikova I, Vizcay-Barrena G, et al. Sporophytic control of pollen meiotic progression is mediated by tapetum expression of ABORTED MICROSPORES[J]. Journal of Experimental Botany, 2022, 73(16): 5543-5558. |
| [26] |
Flores-Tornero M, Anoman AD, Rosa-Téllez S, et al. Lack of phosphoserine phosphatase activity alters pollen and tapetum development in Arabidopsis thaliana[J]. Plant Science, 2015, 235: 81-88.
doi: 10.1016/j.plantsci.2015.03.001 pmid: 25900568 |
| [27] | Chen W, Yu XH, Zhang K, et al. Male Sterile2 encodes a plastid-localized fatty acyl carrier protein reductase required for pollen exine development in Arabidopsis[J]. Plant Physiology, 2011, 157(2): 842-853. |
| [28] | Aarts MG, Keijzer CJ, Stiekema WJ, et al. Molecular characterization of the CER1 gene of arabidopsis involved in epicuticular wax biosynthesis and pollen fertility[J]. The Plant Cell, 1995, 7(12): 2115-2127. |
| [29] | Suzuki T, Narciso JO, Zeng W, et al. KNS4/UPEX1: a type II arabinogalactan β-(1, 3)-galactosyltransferase required for pollen exine development[J]. Plant Physiology, 2017, 173(1): 183-205. |
| [30] | Li JJ, Han SH, Ding XL, et al. Comparative transcriptome analysis between the cytoplasmic male sterile line NJCMS1A and its maintainer NJCMS1B in soybean(Glycine max(L.)Merr.)[J]. PLoS One, 2015, 10(5): e0126771. |
| [31] | Hua MY, Yin WZ, Fernández Gómez J, et al. Barley TAPETAL DEVELOPMENT and FUNCTION1(HvTDF1)gene reveals conserved and unique roles in controlling anther tapetum development in dicot and monocot plants[J]. New Phytologist, 2023, 240(1): 173-190. |
| [32] | Li H, Zhang DB. Biosynthesis of anther cuticle and pollen exine in rice[J]. Plant signaling & behavior, 2010, 5(9): 1121-1123. |
| [33] | Zhao YN, Luo LL, Xu JS, et al. Malate transported from chloroplast to mitochondrion triggers production of ROS and PCD in Arabidopsis thaliana[J]. Cell Research, 2018, 28(4): 448-461. |
| [34] | Liu J, Xia C, Dong HX, et al. Wheat male-sterile 2 reduces ROS levels to inhibit anther development by deactivating ROS modulator 1[J]. Molecular Plant, 2022, 15(9): 1428-1439. |
| [35] | Zhang DD, Liu D, Lv XM, et al. The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen development in Arabidopsis[J]. The Plant Cell, 2014, 26(7): 2939-2961. |
| [36] |
De Veylder L, Beeckman T, Inzé D. The ins and outs of the plant cell cycle[J]. Nature Reviews Molecular Cell Biology, 2007, 8 (8): 655-665.
doi: 10.1038/nrm2227 pmid: 17643126 |
| [37] | Zhang TB, Yuan SH, Liu ZH, et al. Comparative transcriptome analysis reveals hormone signal transduction and sucrose metabolism related genes involved in the regulation of anther dehiscence in photo-thermo-sensitive genic male sterile wheat[J]. Biomolecules, 2022, 12(8): 1149 |
| [38] | He MT, Wang XX, Bu YN, et al. Gibberellin confers to the expression of TaGA-6D and negatively regulates the fertility of wheat with Aegilops juvenalis cytoplasm[J]. Plant Science, 2023: 111771. |
| [39] | Park JH, Halitschke R, Kim HB, et al. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis[J]. The Plant Journal, 2002, 31(1): 1-12. |
| [40] | Li Z, Luo X, Ou Y, et al. JASMONATE-ZIM DOMAIN proteins engage polycomb chromatin modifiers to modulate jasmonate signaling in Arabidopsis[J]. Molecular Plant, 2021, 14(5): 732-747. |
| [41] | Fang YX, Guo DS, Wang Y, et al. Rice transcriptional repressor OsTIE1 controls anther dehiscence and male sterility by regulating JA biosynthesis[J]. The Plant Cell, 2024: koae028. |
| [1] | GAO Meng-meng, ZHAO Tian-yu, JIAO Xin-yue, LIN Chun-jing, GUAN Zhe-yun, DING Xiao-yang, SUN Yan-yan, ZHANG Chun-bao. Comparative Transcriptome Analysis of Cytoplasmic Male Sterile Line and Its Restorer Line in Soybean [J]. Biotechnology Bulletin, 2024, 40(7): 137-149. |
| [2] | BAI Zhi-yuan, XU Fei, YANG Wu, WANG Ming-gui, YANG Yu-hua, ZHANG Hai-ping, ZHANG Rui-jun. Transcriptome Analysis of Fertility Transformation in Weakly Restoring Hybrid F1 of Soybean Cytoplasmic Male Sterility [J]. Biotechnology Bulletin, 2024, 40(6): 134-142. |
| [3] | WU Di, YOU Xiao-feng, ZHENG Yi-zheng, LIN Nan, ZHANG Yan-yan, WEI Yi-cong. Analysis of Endogenous Hormone Regulation Mechanism for Carotenoid Synthesis in Sarcandra glabra [J]. Biotechnology Bulletin, 2024, 40(5): 203-214. |
| [4] | ZHANG Zhen, LI Qing, XU Jing, CHEN Kai-yuan, ZHANG Chun-zhi, ZHU Guang-tao. Construction and Application of Potato Mitochondrial Targeted Expression Vector [J]. Biotechnology Bulletin, 2024, 40(5): 66-73. |
| [5] | GUO Chun, SONG Gui-mei, YAN Yan, DI Peng, WANG Ying-ping. Genome Wide Identification and Expression Analysis of the bZIP Gene Family in Panax quinquefolius [J]. Biotechnology Bulletin, 2024, 40(4): 167-178. |
| [6] | ZHONG Yun, LIN Chun, LIU Zheng-jie, DONG Chen-wen-hua, MAO Zi-chao, LI Xing-yu. Cloning and Prokaryotic Expression Analysis of Asparagus Saponin Synthesis Related Glycosyltransferase Genes [J]. Biotechnology Bulletin, 2024, 40(4): 255-263. |
| [7] | YANG Qi, WEI Zi-di, SONG Juan, TONG Kun, YANG Liu, WANG Jia-han, LIU Hai-yan, LUAN Wei-jiang, MA Xuan. Construction and Transcriptomic Analysis of Rice Histone H1 Triple Mutant [J]. Biotechnology Bulletin, 2024, 40(4): 85-96. |
| [8] | XIE Qian, JIANG Lai, HE Jin, LIU Ling-ling, DING Ming-yue, CHEN Qing-xi. Regulatory Genes Mining Related to Transcriptome Sequencing and Phenolic Metabolism Pathway of Canarium album Fruit with Different Fresh Food Quality [J]. Biotechnology Bulletin, 2024, 40(3): 215-228. |
| [9] | LIN Hong-yan, GUO Xiao-rui, LIU Di, LI Hui, LU Hai. Molecular Mechanism of Transcriptional Factor AtbHLH68 in Regulating Cell Wall Development by Transcriptome Analysis [J]. Biotechnology Bulletin, 2023, 39(9): 105-116. |
| [10] | MIAO Yong-mei, MIAO Cui-ping, YU Qing-cai. Properties of Bacillus subtilis Strain BBs-27 Fermentation Broth and the Inhibition of Lipopeptides Against Fusarium culmorum [J]. Biotechnology Bulletin, 2023, 39(9): 255-267. |
| [11] | FU Yu, JIA Rui-rui, HE He, WANG Liang-gui, YANG Xiu-lian. Growth Differences Among Grafted Seedlings with Two Rootstocks of Catalpa bungei and Comparative Analysis of Transcriptome [J]. Biotechnology Bulletin, 2023, 39(8): 251-261. |
| [12] | KONG De-zhen, DUAN Zhen-yu, WANG Gang, ZHANG Xin, XI Lin-qiao. Physiological Characteristics and Transcriptome Analysis of Sorghum bicolor × S. Sudanense Seedlings Under Salt-alkali Stress [J]. Biotechnology Bulletin, 2023, 39(6): 199-207. |
| [13] | LIU Hui, LU Yang, YE Xi-miao, ZHOU Shuai, LI Jun, TANG Jian-bo, CHEN En-fa. Comparative Transcriptome Analysis of Cadmium Stress Response Induced by Exogenous Sulfur in Tartary Buckwheat [J]. Biotechnology Bulletin, 2023, 39(5): 177-191. |
| [14] | XIE Yang, XING Yu-meng, ZHOU Guo-yan, LIU Mei-yan, YIN Shan-shan, YAN Li-ying. Transcriptome Analysis of Diploid and Autotetraploid in Cucumber Fruit [J]. Biotechnology Bulletin, 2023, 39(3): 152-162. |
| [15] | HU Li-li, LIN Bo-rong, WANG Hong-hong, CHEN Jian-song, LIAO Jin-ling, ZHUO Kan. Transcriptome and Candidate Effectors Analysis of Pratylenchus brachyurus [J]. Biotechnology Bulletin, 2023, 39(3): 254-266. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||