








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (7): 137-149.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0219
Previous Articles Next Articles
GAO Meng-meng1,2,3( ), ZHAO Tian-yu1,2,3, JIAO Xin-yue1,2,3, LIN Chun-jing2,3, GUAN Zhe-yun2,3, DING Xiao-yang2,3, SUN Yan-yan2,3(
), ZHAO Tian-yu1,2,3, JIAO Xin-yue1,2,3, LIN Chun-jing2,3, GUAN Zhe-yun2,3, DING Xiao-yang2,3, SUN Yan-yan2,3( ), ZHANG Chun-bao2,3(
), ZHANG Chun-bao2,3( )
)
Received:2024-03-07
Online:2024-07-26
Published:2024-07-30
Contact:
SUN Yan-yan, ZHANG Chun-bao
E-mail:1757753098@qq.com;15929800833@163.com;cbzhang@cjaas.com
GAO Meng-meng, ZHAO Tian-yu, JIAO Xin-yue, LIN Chun-jing, GUAN Zhe-yun, DING Xiao-yang, SUN Yan-yan, ZHANG Chun-bao. Comparative Transcriptome Analysis of Cytoplasmic Male Sterile Line and Its Restorer Line in Soybean[J]. Biotechnology Bulletin, 2024, 40(7): 137-149.
| 基因名称Gene name | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| Cons4 | GATCAGCAATTATGCACAACG | CCGCCACCATTCAGATTATGT |
| Glyma.01G235800 | CGTGAGAGATGTGGTGACATAC | ACGACCCATCTTCCATTTCC |
| Glyma.08G233900 | TGAGAGATGTGGTGCTTTGG | :GTTTCCCTCCATCCTCCTAAAC |
| Glyma.09G160300 | GATTGGTAGAAGAGCGACTGAA | CGACCGAGAAGATCGATCAAA |
| Glyma.09G176400 | GGTCAACCCGCAGATGTAAT | CGGTCGAATTCCCTGATCTTT |
| Glyma.09G176600 | GTCGCTGCAAACAAGGTTAAG | GGCTGAGAGTGAAAGTGAGAAT |
| Glyma.09G209700 | CAGGACAAGTCTCAACCACAA | CTGTGACATGTCTGGCGTATAA |
| Glyma.10G047150 | CTTCGTGAAGGAGAGTGGAATC | CAAACAGCTCCCGCGTATAA |
| Glyma.13G135000 | CGCGGGTTGTACTCTGATATT | GTTCCACGTTACGATATCTCTCTC |
| Glyma.15G019900 | TGAGCCGGATATTGTGGTTTAC | ACCTCTTCCTCCTCATCTCTTT |
| Glyma.15G091700 | GTTGTATGGGTGTGAGCCTAAT | CCTTCCCTCTCATCTCCCTATAA |
| Glyma.16G034600 | CCGAGGAGTTCAAGTCTTCTTC | CCTGGTTCTTTCGCTGGTAATA |
| Glyma.16G195700 | CTAGGAGATGCCTGTGATTTGT | AAAGCCATGGATCAGAGTAGTG |
| Glyma.16G195900 | CCGGATGCAATTACCCTCAA | ACCCTTGAGCTACGACCTTA |
| Glyma.17G220100 | GTGCCCATAGGAGTCAGAAATAC | CGCGGAGATTCTTCGTTACTT |
| Glyma.18G108202 | GCGGCTTGAGGAAGGTAATAA | TCATGCATCCAGCCTCTTAATC |
Table 1 Information of RT-qPCR primer sequences
| 基因名称Gene name | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| Cons4 | GATCAGCAATTATGCACAACG | CCGCCACCATTCAGATTATGT |
| Glyma.01G235800 | CGTGAGAGATGTGGTGACATAC | ACGACCCATCTTCCATTTCC |
| Glyma.08G233900 | TGAGAGATGTGGTGCTTTGG | :GTTTCCCTCCATCCTCCTAAAC |
| Glyma.09G160300 | GATTGGTAGAAGAGCGACTGAA | CGACCGAGAAGATCGATCAAA |
| Glyma.09G176400 | GGTCAACCCGCAGATGTAAT | CGGTCGAATTCCCTGATCTTT |
| Glyma.09G176600 | GTCGCTGCAAACAAGGTTAAG | GGCTGAGAGTGAAAGTGAGAAT |
| Glyma.09G209700 | CAGGACAAGTCTCAACCACAA | CTGTGACATGTCTGGCGTATAA |
| Glyma.10G047150 | CTTCGTGAAGGAGAGTGGAATC | CAAACAGCTCCCGCGTATAA |
| Glyma.13G135000 | CGCGGGTTGTACTCTGATATT | GTTCCACGTTACGATATCTCTCTC |
| Glyma.15G019900 | TGAGCCGGATATTGTGGTTTAC | ACCTCTTCCTCCTCATCTCTTT |
| Glyma.15G091700 | GTTGTATGGGTGTGAGCCTAAT | CCTTCCCTCTCATCTCCCTATAA |
| Glyma.16G034600 | CCGAGGAGTTCAAGTCTTCTTC | CCTGGTTCTTTCGCTGGTAATA |
| Glyma.16G195700 | CTAGGAGATGCCTGTGATTTGT | AAAGCCATGGATCAGAGTAGTG |
| Glyma.16G195900 | CCGGATGCAATTACCCTCAA | ACCCTTGAGCTACGACCTTA |
| Glyma.17G220100 | GTGCCCATAGGAGTCAGAAATAC | CGCGGAGATTCTTCGTTACTT |
| Glyma.18G108202 | GCGGCTTGAGGAAGGTAATAA | TCATGCATCCAGCCTCTTAATC |
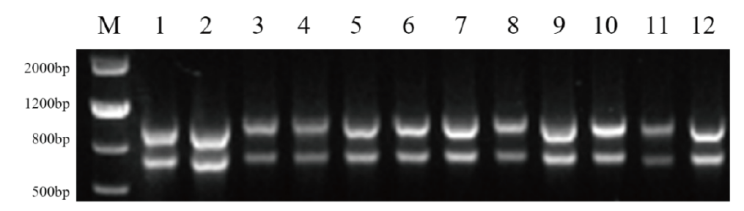
Fig. 2 Result of total RNA in flower bud by agarose gel electrophoresis M: Maker Ⅲ. Lane 1-3: FLB_1 - FLB_3; respectively. Lane 4-6: FSB_1 - FSB_3; respectively. Lane 7-9: SLB_1 -SLB_3; respectively. Lane 10-12: SSB_1 - SSB_3; respectively
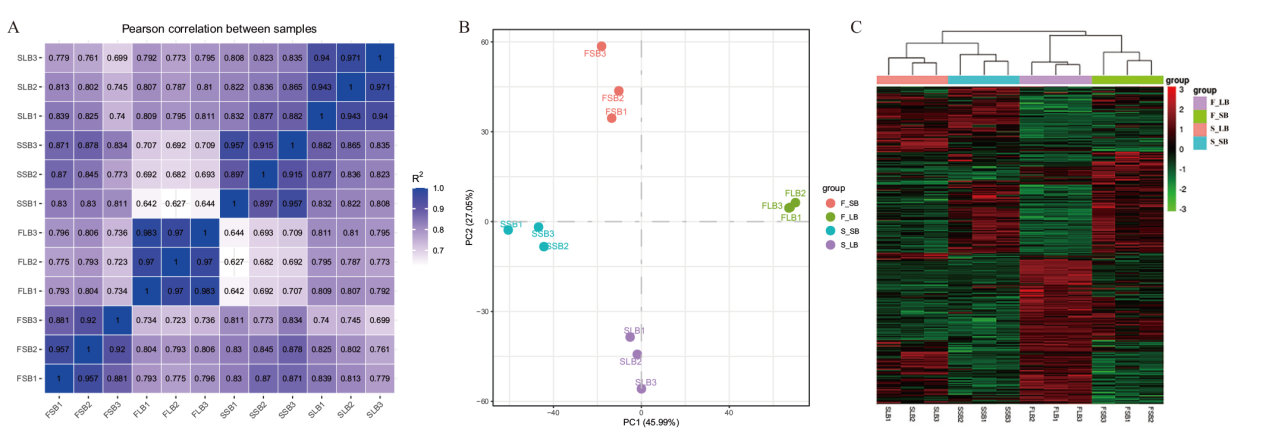
Fig. 3 Correlation analysis among samples A: Pearson correlation analysis of fertile and sterile samples at different developmental stages. B: PCA analysis of gene expression levels in fertile and sterile samples at different developmental stages. C: Heat map of gene expressions among samples
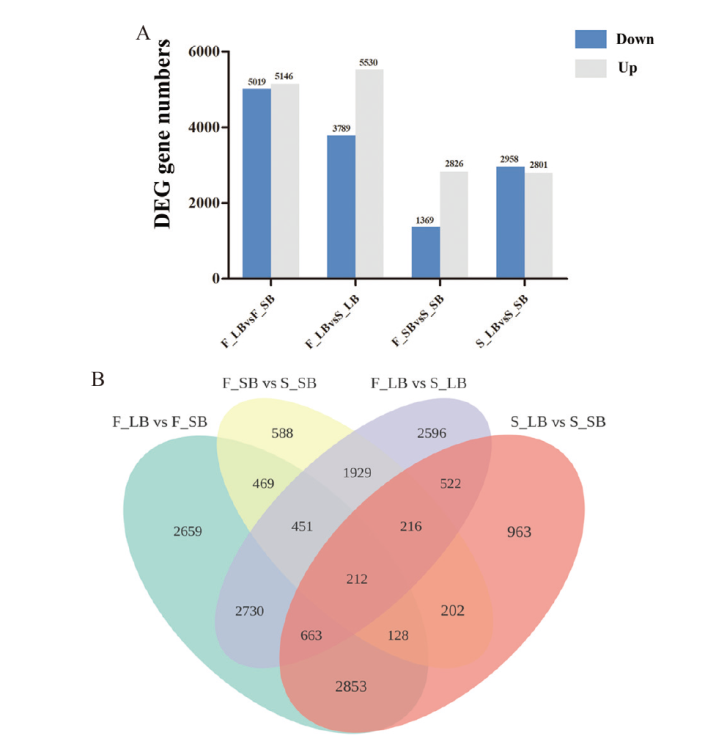
Fig. 4 Statistics and analysis of differentially expressed genes A: Statistics of differentially expressed genes. B: Venn diagram of differentially expressed genes
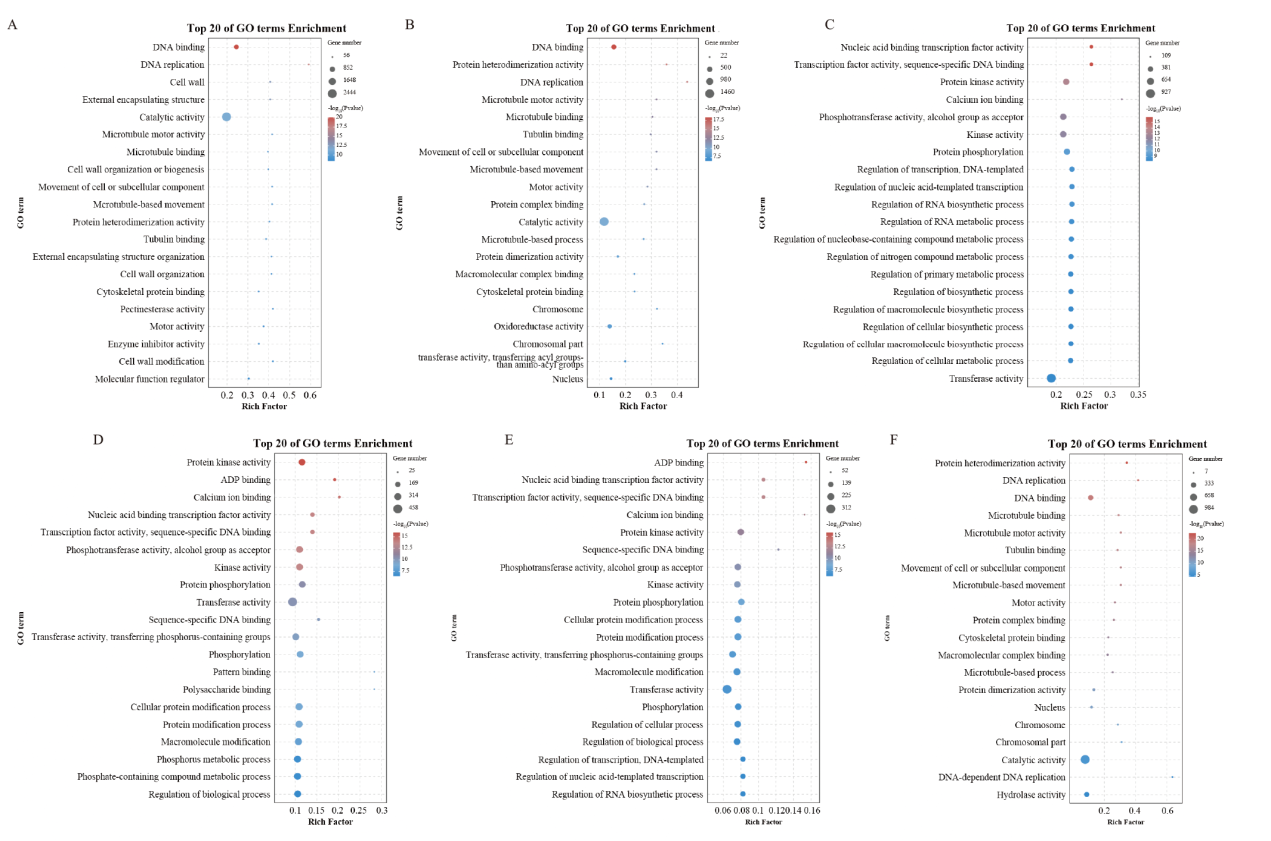
Fig. 5 Rich distribution point diagram for GO A:F_LB vs F_SB;B:S_LB vs S_SB;C:F_LB vs S_LB;D:F_SB vs S_SB;E:F_LB vs S_LB和F_SB vs S_SB;F:F_LB vs F_SB和S_LB vs S_SB, the same below
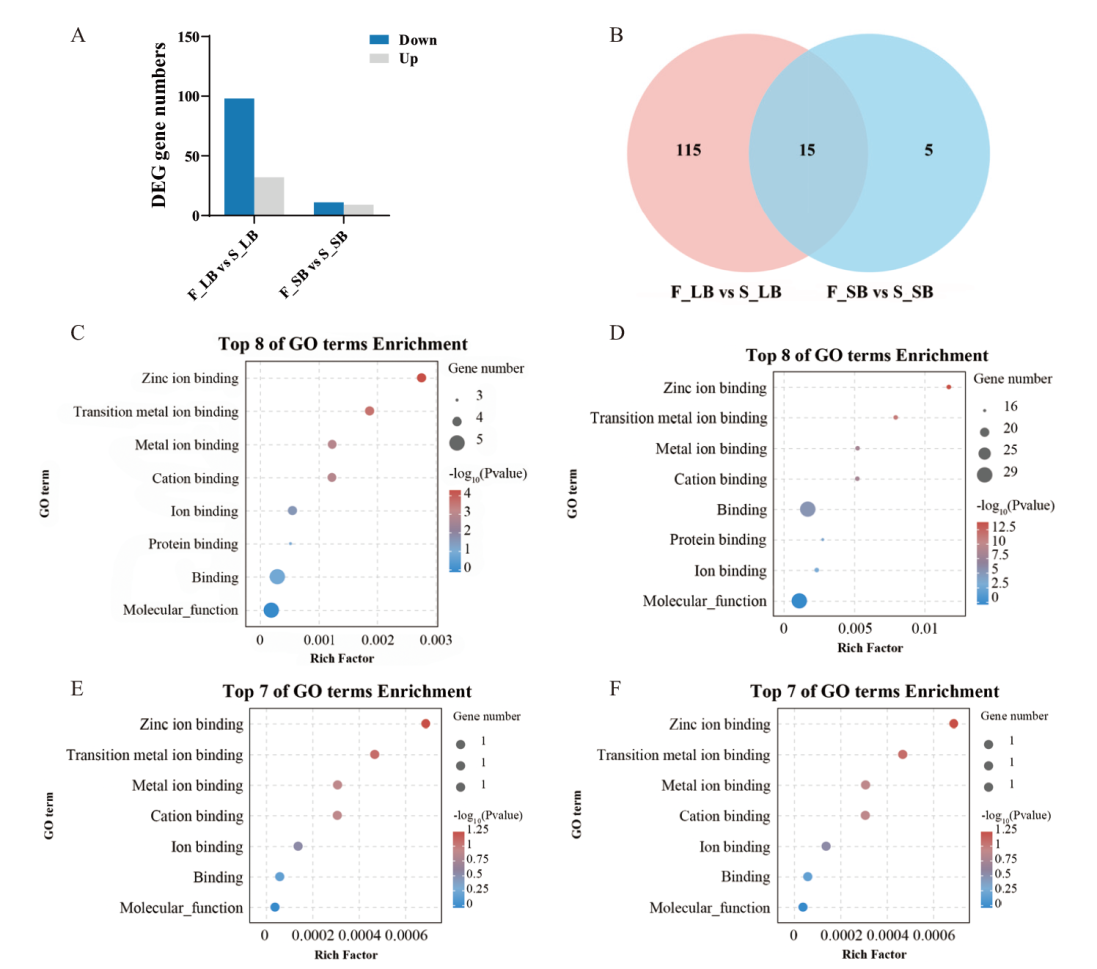
Fig. 7 Statistical analysis of differentially expressed genes encoding PPR proteins A: Statistics of differentially expressed genes encoding PPR protein. B: Venn diagram of differentially expressed genes encoding PPR protein. C: Map of GO enrichment of up-regulated DEGs in F_LB vs S_LB. D: Map of GO enrichment of down-regulated DEGs in F_LB vs S_LB. E: Map of GO enrichment of up-regulated DEGs in F_SB vs S_SB. F: Map of GO enrichment of down-regulated DEGs in F_SB vs S_SB
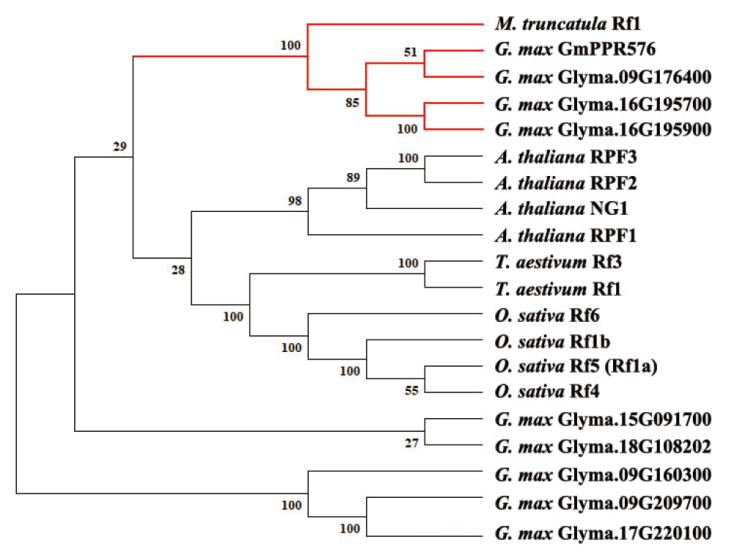
Fig. 9 Phylogenetic analysis of candidate genes encoding PPR proteins The red branches indicate PPR proteins in leguminous plants. Phylogenetic analysis was constructed using Maximum-Likelihood, and Bootstrap values of 500 were used
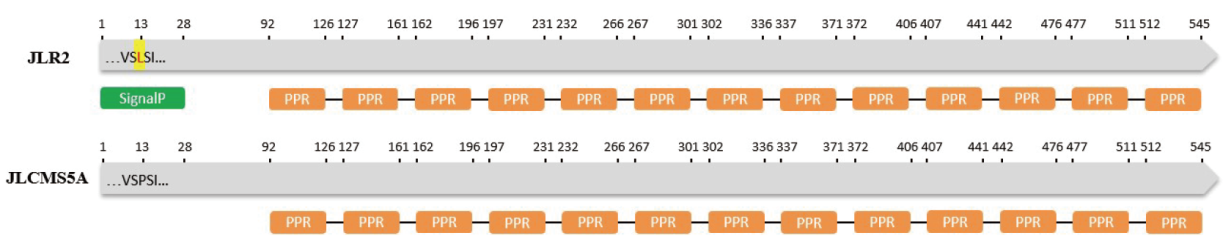
Fig. 10 Structure prediction for Glyma.09G176400 encoded protein Green graphic indicates signal peptides. The orange graphics indicate pentatricopeptide repeat. The yellow graphic indicate amino acid mutation site
| [1] | 张春宝, 孙妍妍, 赵丽梅. 大豆细胞质雄性不育遗传基础与育种应用[J]. 植物遗传资源学报, 2024, 25(6): 857-869. |
| Zhang CB, Sun YY, Zhao LM. Genetic basis and breeding application of cytoplasmic male sterility in soybean[J]. J Plant Genet Resour, 2024, 25(6): 857-869. | |
| [2] |
Chen LT, Liu YG. Male sterility and fertility restoration in crops[J]. Annu Rev Plant Biol, 2014, 65: 579-606.
doi: 10.1146/annurev-arplant-050213-040119 pmid: 24313845 |
| [3] | Levings CS. Thoughts on cytoplasmic male sterility in cms-T maize[J]. Plant Cell, 1993, 5(10): 1285-1290. |
| [4] | Grelon M, Budar F, Bonhomme S, et al. Ogura cytoplasmic male-sterility(CMS)-associated orf138 is translated into a mitochondrial membrane polypeptide in male-sterile Brassica cybrids[J]. Mol Gen Genet, 1994, 243(5): 540-547. |
| [5] | Schnable P. The molecular basis of cytoplasmic male sterility and fertility restoration[J]. Trends Plant Sci, 1998, 3(5): 175-180. |
| [6] |
Balk J, Leaver CJ. The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release[J]. Plant Cell, 2001, 13(8): 1803-1818.
doi: 10.1105/tpc.010116 pmid: 11487694 |
| [7] | Qin XE, Tian SK, Zhang WL, et al. The main restorer Rf3 of maize S type cytoplasmic male sterility encodes a PPR protein that functions in reduction of the transcripts of orf355[J]. Mol Plant, 2021, 14(12): 1961-1964. |
| [8] | Jiang HC, Lu Q, Qiu SQ, et al. Fujian cytoplasmic male sterility and the fertility restorer gene OsRf19 provide a promising breeding system for hybrid rice[J]. Proc Natl Acad Sci USA, 2022, 119(34): e2208759119. |
| [9] | Lin YN, Yang HL, Liu HM, et al. A P-type pentatricopeptide repeat protein ZmRF5 promotes 5' region partial cleavages of atp6c transcripts to restore the fertility of CMS-C maize by recruiting a splicing factor[J]. Plant Biotechnol J, 2023: 1-13. |
| [10] |
Schmitz-Linneweber C, Williams-Carrier R, Barkan A. RNA immunoprecipitation and microarray analysis show a chloroplast Pentatricopeptide repeat protein to be associated with the 5' region of mRNAs whose translation it activates[J]. Plant Cell, 2005, 17(10): 2791-2804.
doi: 10.1105/tpc.105.034454 pmid: 16141451 |
| [11] |
Beick S, Schmitz-Linneweber C, Williams-Carrier R, et al. The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts[J]. Mol Cell Biol, 2008, 28(17): 5337-5347.
doi: 10.1128/MCB.00563-08 pmid: 18591259 |
| [12] |
Pfalz J, Bayraktar OA, Prikryl J, et al. Site-specific binding of a PPR protein defines and stabilizes 5' and 3' mRNA termini in chloroplasts[J]. EMBO J, 2009, 28(14): 2042-2052.
doi: 10.1038/emboj.2009.121 pmid: 19424177 |
| [13] | Kotera E, Tasaka M, Shikanai T. A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts[J]. Nature, 2005, 433(7023): 326-330. |
| [14] | Okuda K, Hammani K, Tanz SK, et al. The pentatricopeptide repeat protein OTP82 is required for RNA editing of plastid ndhB and ndhG transcripts[J]. Plant J, 2010, 61(2): 339-349. |
| [15] |
Robbins JC, Heller WP, Hanson MR. A comparative genomics approach identifies a PPR-DYW protein that is essential for C-to-U editing of the Arabidopsis chloroplast accD transcript[J]. RNA, 2009, 15(6): 1142-1153.
doi: 10.1261/rna.1533909 pmid: 19395655 |
| [16] |
Gillman JD, Bentolila S, Hanson MR. The petunia restorer of fertility protein is part of a large mitochondrial complex that interacts with transcripts of the CMS-associated locus[J]. Plant J, 2007, 49(2): 217-227.
doi: 10.1111/j.1365-313X.2006.02953.x pmid: 17156410 |
| [17] |
Brown GG, Formanová N, Jin H, et al. The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats[J]. Plant J, 2003, 35(2): 262-272.
doi: 10.1046/j.1365-313x.2003.01799.x pmid: 12848830 |
| [18] |
Liu Z, Yang ZH, Wang X, et al. A mitochondria-targeted PPR protein restores pol cytoplasmic male sterility by reducing orf224 transcript levels in oilseed rape[J]. Mol Plant, 2016, 9(7): 1082-1084.
doi: 10.1016/j.molp.2016.04.004 pmid: 27102212 |
| [19] | Liu Z, Dong FM, Wang X, et al. A pentatricopeptide repeat protein restores nap cytoplasmic male sterility in Brassica napus[J]. J Exp Bot, 2017, 68(15): 4115-4123. |
| [20] | Wang HD, Xiao Q, Wei C, et al. A mitochondria-localized pentatricopeptide repeat protein is required to restore hau cytoplasmic male sterility in Brassica napus[J]. Theor Appl Genet, 2021, 134(5): 1377-1386. |
| [21] | Komori T, Ohta S, Murai N, et al. Map-based cloning of a fertility restorer gene, Rf-1, in rice(Oryza sativa L.)[J]. Plant J, 2004, 37(3): 315-325. |
| [22] | Hu J, Wang K, Huang WC, et al. The rice pentatricopeptide repeat protein RF5 restores fertility in Hong-Lian cytoplasmic male-sterile lines via a complex with the glycine-rich protein GRP162[J]. Plant Cell, 2012, 24(1): 109-122. |
| [23] |
Tang HW, Luo DP, Zhou DG, et al. The rice restorer Rf4 for wild-abortive cytoplasmic male sterility encodes a mitochondrial-localized PPR protein that functions in reduction of WA352 transcripts[J]. Mol Plant, 2014, 7(9): 1497-1500.
doi: S1674-2052(14)60953-9 pmid: 24728538 |
| [24] |
Huang WC, Yu CC, Hu J, et al. Pentatricopeptide-repeat family protein RF6 functions with hexokinase 6 to rescue rice cytoplasmic male sterility[J]. Proc Natl Acad Sci USA, 2015, 112(48): 14984-14989.
doi: 10.1073/pnas.1511748112 pmid: 26578814 |
| [25] | Melonek J, Duarte J, Martin J, et al. The genetic basis of cytoplasmic male sterility and fertility restoration in wheat[J]. Nat Commun, 2021, 12(1): 1036. |
| [26] |
Wang TL, He TT, Ding XL, et al. Confirmation of GmPPR576 as a fertility restorer gene of cytoplasmic male sterility in soybean[J]. J Exp Bot, 2021, 72(22): 7729-7742.
doi: 10.1093/jxb/erab382 pmid: 34397079 |
| [27] |
杨绪磊, 郭凤兰, 高萌萌, 等. 大豆CMS-RN型不育系育性恢复基因GmRf1的初步鉴定及其分子标记开发[J]. 植物遗传资源学报, 2023, 24(4): 1186-1193.
doi: 10.13430/j.cnki.jpgr.20221124001 |
| Yang XL, Guo FL, Gao MM, et al. Preliminary identification and molecular marker development of the restorer-of-fertility gene GmRf1 of CMS-RN type sterile lines in soybean[J]. J Plant Genet Resour, 2023, 24(4): 1186-1193. | |
| [28] |
Liu YC, Du HL, Li PC, et al. Pan-genome of wild and cultivated soybeans[J]. Cell, 2020, 182(1): 162-176.e13.
doi: S0092-8674(20)30618-8 pmid: 32553274 |
| [29] |
Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features[J]. Bioinformatics, 2014, 30(7): 923-930.
doi: 10.1093/bioinformatics/btt656 pmid: 24227677 |
| [30] |
Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation[J]. Nat Biotechnol, 2010, 28(5): 511-515.
doi: 10.1038/nbt.1621 pmid: 20436464 |
| [31] | Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2[J]. Genome Biol, 2014, 15(12): 550. |
| [32] |
Gaborieau L, Brown GG, Mireau H. The propensity of pentatricopeptide repeat genes to evolve into restorers of cytoplasmic male sterility[J]. Front Plant Sci, 2016, 7: 1816.
doi: 10.3389/fpls.2016.01816 pmid: 27999582 |
| [33] | Li JJ, Yang SP, Gai JY. Transcriptome comparative analysis between the cytoplasmic male sterile line and fertile line in soybean(Glycine max(L.)Merr.)[J]. Genes Genom, 2017, 39(10): 1117-1127. |
| [34] | Li JJ, Han SH, Ding XL, et al. Comparative transcriptome analysis between the cytoplasmic male sterile line NJCMS1A and its maintainer NJCMS1B in soybean(Glycine max(L.)Merr.)[J]. PLoS One, 2015, 10(5): e0126771. |
| [35] | 赵颖. 联合RNA-seq和iTRAQ技术挖掘大豆细胞质雄性不育相关基因[D]. 通辽: 内蒙古民族大学, 2023. |
| Zhao Y. Combined RNA-seq and iTRAQ technology to mine genes related to cytoplasmic male sterility in soybean[D]. Tongliao: Inner Mongolia University for the Nationalities, 2023. | |
| [36] | 姜童, 付翔, 王辉, 等. 簇生朝天椒雄性不育系及保持系的花器官及生理特性研究[J]. 北方园艺, 2018(11): 22-26. |
| Jiang T, Fu X, Wang H, et al. Flower morphological and physiological characters between male sterile line and maintainer line in cluster pepper[J]. North Hortic, 2018(11): 22-26. | |
| [37] | 鲁美宏, 孙万仓, 孔德晶, 等. 白菜型冬油菜不育系LRCMS花器生理生化特性及其雄蕊发育特征研究[J]. 西北植物学报, 2014, 34(3): 509-515. |
| Lu MH, Sun WC, Kong DJ, et al. Physiobiochemical characteristics and stamen development characteristics of LRCMS flower in winter rapeseed(Brassica campestris)[J]. Acta Bot Boreali Occidentalia Sin, 2014, 34(3): 509-515. | |
| [38] |
白志元, 杨玉花, 张瑞军. 不同恢复型大豆细胞质雄性不育杂种F1的转录组分析[J]. 植物遗传资源学报, 2022, 23(6): 1847-1855.
doi: 10.13430/j.cnki.jpgr.20220510001 |
| Bai ZY, Yang YH, Zhang RJ. Transcriptomic analysis of soybean cytoplasmic male sterile F1 hybrids from pollination with different restorer types[J]. J Plant Genet Resour, 2022, 23(6): 1847-1855. | |
| [39] | Barkan A, Rojas M, Fujii S, et al. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins[J]. PLoS Genet, 2012, 8(8): e1002910. |
| [40] | Kazama T, Toriyama K. A fertility restorer gene, Rf4, widely used for hybrid rice breeding encodes a pentatricopeptide repeat protein[J]. Rice, 2014, 7(1): 28. |
| [41] |
Wang ZH, Zou YJ, Li XY, et al. Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing[J]. Plant Cell, 2006, 18(3): 676-687.
doi: 10.1105/tpc.105.038240 pmid: 16489123 |
| [42] | Wang CD, Lezhneva L, Arnal N, et al. The radish Ogura fertility restorer impedes translation elongation along its cognate CMS-causing mRNA[J]. Proc Natl Acad Sci USA, 2021, 118(35): e2105274118. |
| [1] | WANG Fang, YU Lu, QI Ze-zheng, ZHOU Chang-jun, YU Ji-dong. Screening and Biocontrol Effect of Antagonistic Bacteria against Soybean Root Rot [J]. Biotechnology Bulletin, 2024, 40(7): 216-225. |
| [2] | BAI Zhi-yuan, XU Fei, YANG Wu, WANG Ming-gui, YANG Yu-hua, ZHANG Hai-ping, ZHANG Rui-jun. Transcriptome Analysis of Fertility Transformation in Weakly Restoring Hybrid F1 of Soybean Cytoplasmic Male Sterility [J]. Biotechnology Bulletin, 2024, 40(6): 134-142. |
| [3] | ZHANG Zhen, LI Qing, XU Jing, CHEN Kai-yuan, ZHANG Chun-zhi, ZHU Guang-tao. Construction and Application of Potato Mitochondrial Targeted Expression Vector [J]. Biotechnology Bulletin, 2024, 40(5): 66-73. |
| [4] | LOU Yin, GAO Hao-jun, WANG Xi, NIU Jing-ping, WANG Min, DU Wei-jun, YUE Ai-qin. Identification and Expression Pattern Analysis of GmHMGS Gene in Soybean [J]. Biotechnology Bulletin, 2024, 40(4): 110-121. |
| [5] | YANG Qi, WEI Zi-di, SONG Juan, TONG Kun, YANG Liu, WANG Jia-han, LIU Hai-yan, LUAN Wei-jiang, MA Xuan. Construction and Transcriptomic Analysis of Rice Histone H1 Triple Mutant [J]. Biotechnology Bulletin, 2024, 40(4): 85-96. |
| [6] | LOU Hui, ZHU Jin-cheng, YANG Yang, ZHANG Wei. Effects of Root Exudates in Resistant and Susceptible Varieties of Cotton on the Growths and Gene Expressions of Fusarium oxysporum [J]. Biotechnology Bulletin, 2023, 39(9): 156-167. |
| [7] | FU Yu, JIA Rui-rui, HE He, WANG Liang-gui, YANG Xiu-lian. Growth Differences Among Grafted Seedlings with Two Rootstocks of Catalpa bungei and Comparative Analysis of Transcriptome [J]. Biotechnology Bulletin, 2023, 39(8): 251-261. |
| [8] | WANG Shuai, FENG Yu-mei, BAI Miao, DU Wei-jun, YUE Ai-qin. Functional Analysis of Soybean Gene GmHMGR Responding to Exogenous Hormones and Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(7): 131-142. |
| [9] | LI Wen-chen, LIU Xin, KANG Yue, LI Wei, QI Ze-zheng, YU Lu, WANG Fang. Optimization and Application of Tobacco Rattle Virus-induced Gene Silencing System in Soybean [J]. Biotechnology Bulletin, 2023, 39(7): 143-150. |
| [10] | YANG Yang, ZHU Jin-cheng, LOU Hui, HAN Ze-gang, ZHANG Wei. Transcriptome Analysis of Interaction Between Gossypium barbadense and Fusarium oxysporum f. sp. vasinfectum [J]. Biotechnology Bulletin, 2023, 39(6): 259-273. |
| [11] | ZHAI Ying, LI Ming-yang, ZHANG Jun, ZHAO Xu, YU Hai-wei, LI Shan-shan, ZHAO Yan, ZHANG Mei-juan, SUN Tian-guo. Heterologous Expression of Soybean Transcription Factor GmNF-YA19 Improves Drought Resistance of Transgenic Tobacco [J]. Biotechnology Bulletin, 2023, 39(5): 224-232. |
| [12] | HOU Xiao-yuan, CHE Zheng-zheng, LI Heng-jing, DU Chong-yu, XU Qian, WANG Qun-qing. Construction of the Soybean Membrane System cDNA Library and Interaction Proteins Screening for Effector PsAvr3a [J]. Biotechnology Bulletin, 2023, 39(4): 268-276. |
| [13] | YANG Chun-hong, DONG Lu, CHEN Lin, SONG Li. Characterization of Soybean VAS1 Gene Family and Its Involvement in Lateral Root Development [J]. Biotechnology Bulletin, 2023, 39(3): 133-142. |
| [14] | XIE Yang, XING Yu-meng, ZHOU Guo-yan, LIU Mei-yan, YIN Shan-shan, YAN Li-ying. Transcriptome Analysis of Diploid and Autotetraploid in Cucumber Fruit [J]. Biotechnology Bulletin, 2023, 39(3): 152-162. |
| [15] | CHEN Yi-bo, YANG Wan-ming, YUE Ai-qin, WANG Li-xiang, DU Wei-jun, WANG Min. Construction of Soybean Genetic Map Based on SLAF Markers and QTL Mapping Analysis of Salt Tolerance at Seedling Stage [J]. Biotechnology Bulletin, 2023, 39(2): 70-79. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||