








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (3): 294-307.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0448
CHE Jian-mei( ), ZHENG Xue-fang, WANG Jie-ping, CHEN Yan-ping, CHEN Bing-xing, LIU Bo(
), ZHENG Xue-fang, WANG Jie-ping, CHEN Yan-ping, CHEN Bing-xing, LIU Bo( )
)
Received:2024-05-14
Online:2025-03-26
Published:2025-03-20
Contact:
LIU Bo
E-mail:chejm2002@163.com;fzliubo@163.com
CHE Jian-mei, ZHENG Xue-fang, WANG Jie-ping, CHEN Yan-ping, CHEN Bing-xing, LIU Bo. Screening, Identification and Whole Genome Analysis of a Cellulase Producing Strain[J]. Biotechnology Bulletin, 2025, 41(3): 294-307.
菌株 Strains | 菌落直径 Colony diameter (d)/mm | 透明圈直径 Diameter of a transparent circle (D)/mm | D/d |
|---|---|---|---|
| FJAT-16079 | 7.41±0.48 | 16.60±5.55 | 2.30 |
| FJAT-16350 | 6.94±4.37 | 16.46±6.27 | 2.99 |
| FJAT-16989 | 3.91±0.22 | 7.02±0.70 | 1.81 |
| FJAT-25102 | 3.44±0.23 | 14.59±0.14 | 4.26 |
| FJAT-17136 | 9.31±1.03 | 11.79±0.37 | 1.28 |
| FJAT-10006 | 3.03±1.01 | 8.85±3.61 | 2.84 |
| FJAT-20218 | 2.16±0.44 | 8.70±1.93 | 4.02 |
| FJAT-22285 | 6.99±0.59 | 9.97±0.18 | 1.43 |
| FJAT-16396 | 6.19±1.26 | 13.64±7.30 | 2.05 |
| FJAT-20520 | 4.36±0.75 | 9.79±0.07 | 2.31 |
| FJAT-20398 | 2.93±0.42 | 11.46±1.29 | 4.06 |
| FJAT-16034 | 6.93±0.25 | 13.46±0.02 | 1.95 |
| FJAT-16517 | 14.72±0.73 | 21.16±0.20 | 1.44 |
| FJAT-26369 | 10.28±0.37 | 12.54±0.24 | 1.22 |
| FJAT-41640 | 6.96±1.12 | 10.00±0.16 | 1.47 |
| FJAT-25102 | 3.74±0.15 | 14.71±0.43 | 3.94 |
| FJAT-22275 | 2.74±0.54 | 7.88±0.44 | 2.96 |
| FJAT-4326 | 9.05±2.12 | 20.03±0.82 | 2.32 |
| FJAT-41635 | 9.22±1.03 | 21.28±1.35 | 2.32 |
Table 1 Screening of cellulose degrading strains
菌株 Strains | 菌落直径 Colony diameter (d)/mm | 透明圈直径 Diameter of a transparent circle (D)/mm | D/d |
|---|---|---|---|
| FJAT-16079 | 7.41±0.48 | 16.60±5.55 | 2.30 |
| FJAT-16350 | 6.94±4.37 | 16.46±6.27 | 2.99 |
| FJAT-16989 | 3.91±0.22 | 7.02±0.70 | 1.81 |
| FJAT-25102 | 3.44±0.23 | 14.59±0.14 | 4.26 |
| FJAT-17136 | 9.31±1.03 | 11.79±0.37 | 1.28 |
| FJAT-10006 | 3.03±1.01 | 8.85±3.61 | 2.84 |
| FJAT-20218 | 2.16±0.44 | 8.70±1.93 | 4.02 |
| FJAT-22285 | 6.99±0.59 | 9.97±0.18 | 1.43 |
| FJAT-16396 | 6.19±1.26 | 13.64±7.30 | 2.05 |
| FJAT-20520 | 4.36±0.75 | 9.79±0.07 | 2.31 |
| FJAT-20398 | 2.93±0.42 | 11.46±1.29 | 4.06 |
| FJAT-16034 | 6.93±0.25 | 13.46±0.02 | 1.95 |
| FJAT-16517 | 14.72±0.73 | 21.16±0.20 | 1.44 |
| FJAT-26369 | 10.28±0.37 | 12.54±0.24 | 1.22 |
| FJAT-41640 | 6.96±1.12 | 10.00±0.16 | 1.47 |
| FJAT-25102 | 3.74±0.15 | 14.71±0.43 | 3.94 |
| FJAT-22275 | 2.74±0.54 | 7.88±0.44 | 2.96 |
| FJAT-4326 | 9.05±2.12 | 20.03±0.82 | 2.32 |
| FJAT-41635 | 9.22±1.03 | 21.28±1.35 | 2.32 |
项目 Item | 结果 Results | 项目 Item | 结果 Results |
|---|---|---|---|
| 精氨酸水解酶Arginine hydrolase | - | 苦杏仁苷Amygdalin | - |
| 苯丙氨酸脱氨酶Phenylalanine deaminase | - | 甘露醇Mannitol | - |
| VP反应Voges-Proskauer test | + | 肌醇Inositol | - |
| 赖氨酸脱羧酶Lysine decarboxylase | - | 山梨醇Sorbitol | - |
| 鸟氨酸脱羧酶Ornithine decarboxylase | - | 密二糖Melibiose | - |
| 柠檬酸盐利用Sodium critrate | + | 葡萄糖Glucose | - |
| 明胶液化Gelatin hydrolysis | + | 阿拉伯糖 | - |
| β-半乳糖苷酶 O-Nitrophenyl-β- | - | 鼠李糖Rhamnose | - |
| 蔗糖Sucrose | - | ||
| 尿素酶Urease | + | 吲哚Indole | - |
| 硫化氢Hydrogen sulfide | - |
Table 2 API 20E results of strain FJAT-25102
项目 Item | 结果 Results | 项目 Item | 结果 Results |
|---|---|---|---|
| 精氨酸水解酶Arginine hydrolase | - | 苦杏仁苷Amygdalin | - |
| 苯丙氨酸脱氨酶Phenylalanine deaminase | - | 甘露醇Mannitol | - |
| VP反应Voges-Proskauer test | + | 肌醇Inositol | - |
| 赖氨酸脱羧酶Lysine decarboxylase | - | 山梨醇Sorbitol | - |
| 鸟氨酸脱羧酶Ornithine decarboxylase | - | 密二糖Melibiose | - |
| 柠檬酸盐利用Sodium critrate | + | 葡萄糖Glucose | - |
| 明胶液化Gelatin hydrolysis | + | 阿拉伯糖 | - |
| β-半乳糖苷酶 O-Nitrophenyl-β- | - | 鼠李糖Rhamnose | - |
| 蔗糖Sucrose | - | ||
| 尿素酶Urease | + | 吲哚Indole | - |
| 硫化氢Hydrogen sulfide | - |
项目 Item | 结果 Results | 项目 Item | 结果 Results |
|---|---|---|---|
| 甘油Glycerol | - | 柳醇Saligenin | - |
| - | 纤维二糖Cellobiose | - | |
| - | 麦芽糖Maltose | - | |
| - | - | ||
| - | - | ||
| β-甲基- | - | - | |
| 赤癣醇Erythritol | - | - | |
| 阿东醇Adonitol | - | - | |
| 果糖Fructose | - | - | |
| 核糖Ribose | - | 龙胆二糖Gentiobiose | - |
| 甘露醇Mannitol | - | 肝糖Glycogen | - |
| 甘露糖Mannose | - | 菊糖Inulin | - |
| 半乳糖Galactose | - | 蜜二糖 | - |
| 半乳糖醇Dulcitol | - | 海藻糖Trehalose | - |
| 肌醇Inositol | - | 蔗糖Sucrose | - |
| 山梨醇Sorbitol | - | 棉子糖Raffinose | - |
| 山梨糖Sorbose | - | 乳糖Lactose | - |
| 葡萄糖Glucose | - | 松三糖Melizitose | - |
| 鼠李糖Rhamnose | - | 木糖醇Xylitol | - |
| 七叶灵Esculin | + | - | |
| 熊果苷Arbutin | - | 5-酮基-葡萄糖酸盐5-keto- | - |
| α-甲基- | - | 2-酮基-葡萄糖酸盐2-keto- | - |
| α-甲基- | - | 葡萄糖酸盐Gluconate | - |
| N-乙酰-葡糖胺N-acetylglucosamine | - | 苦杏仁苷Amygdalin | - |
| 淀粉Starch | - |
Table 3 API 50CH experimental results of strain FJAT-25102
项目 Item | 结果 Results | 项目 Item | 结果 Results |
|---|---|---|---|
| 甘油Glycerol | - | 柳醇Saligenin | - |
| - | 纤维二糖Cellobiose | - | |
| - | 麦芽糖Maltose | - | |
| - | - | ||
| - | - | ||
| β-甲基- | - | - | |
| 赤癣醇Erythritol | - | - | |
| 阿东醇Adonitol | - | - | |
| 果糖Fructose | - | - | |
| 核糖Ribose | - | 龙胆二糖Gentiobiose | - |
| 甘露醇Mannitol | - | 肝糖Glycogen | - |
| 甘露糖Mannose | - | 菊糖Inulin | - |
| 半乳糖Galactose | - | 蜜二糖 | - |
| 半乳糖醇Dulcitol | - | 海藻糖Trehalose | - |
| 肌醇Inositol | - | 蔗糖Sucrose | - |
| 山梨醇Sorbitol | - | 棉子糖Raffinose | - |
| 山梨糖Sorbose | - | 乳糖Lactose | - |
| 葡萄糖Glucose | - | 松三糖Melizitose | - |
| 鼠李糖Rhamnose | - | 木糖醇Xylitol | - |
| 七叶灵Esculin | + | - | |
| 熊果苷Arbutin | - | 5-酮基-葡萄糖酸盐5-keto- | - |
| α-甲基- | - | 2-酮基-葡萄糖酸盐2-keto- | - |
| α-甲基- | - | 葡萄糖酸盐Gluconate | - |
| N-乙酰-葡糖胺N-acetylglucosamine | - | 苦杏仁苷Amygdalin | - |
| 淀粉Starch | - |

Fig. 4 Phylogenetic trees of the strain FJAT-25102 based on the 16S rRNA gene sequencesThe serial number in brackets is the GenBank accession number of the strain. The value of 0.50 indicates the sequence deviation value. The number of internal nodes is support value, which indicates the reliability of the branch structure. The same below
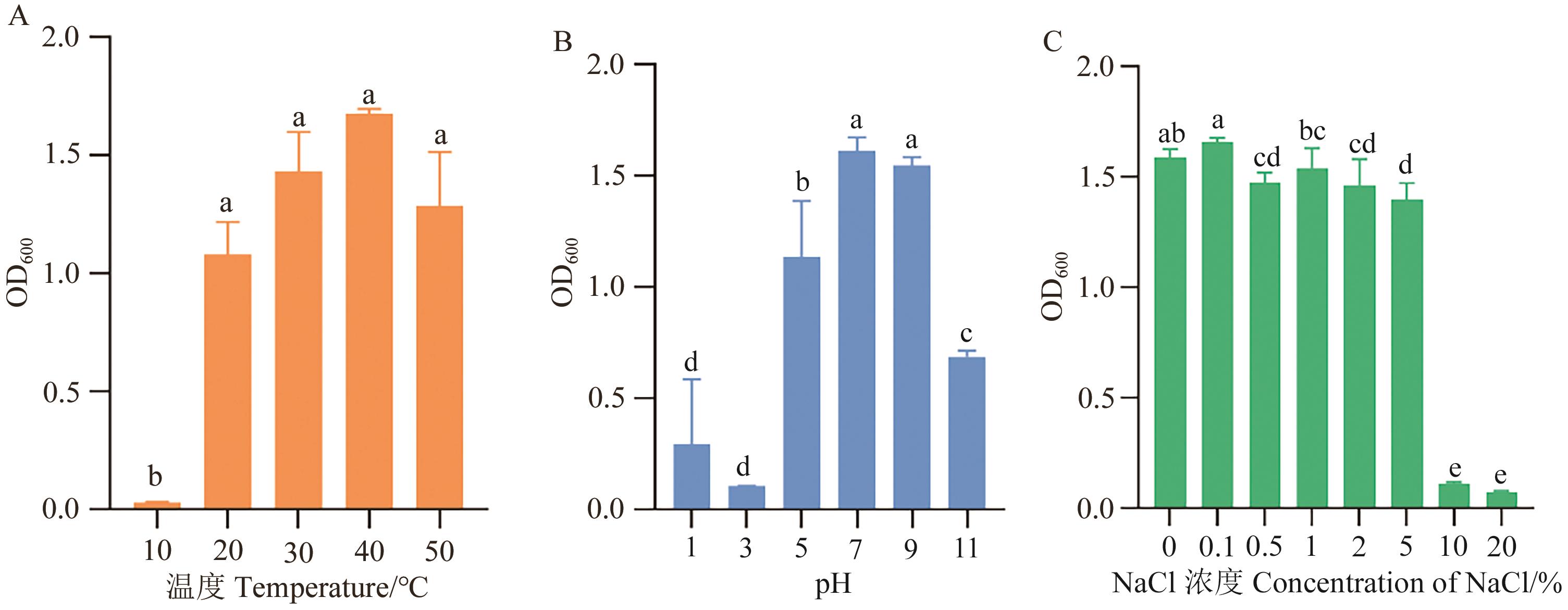
Fig. 6 Effect of different conditions on growth of strain FJAT-25102Different letters indicate significant difference at P<0.05 level among different treatments. The same below

Fig. 7 Circular genome circle map of strain FJAT-25102The outermost circle is the genome size. The second and third circles are the CDS on the positive and negative strands. The functional classifications of COGs of different CDS are distinguished by different colors. The fourth circle is rRNA and tRNA. The fifth circle is the GC content. The outward red indicates the GC content is higher than the genome level, and the inward blue indicates that the GC content is lower than the genome level. The innermost circle is GC-skew value
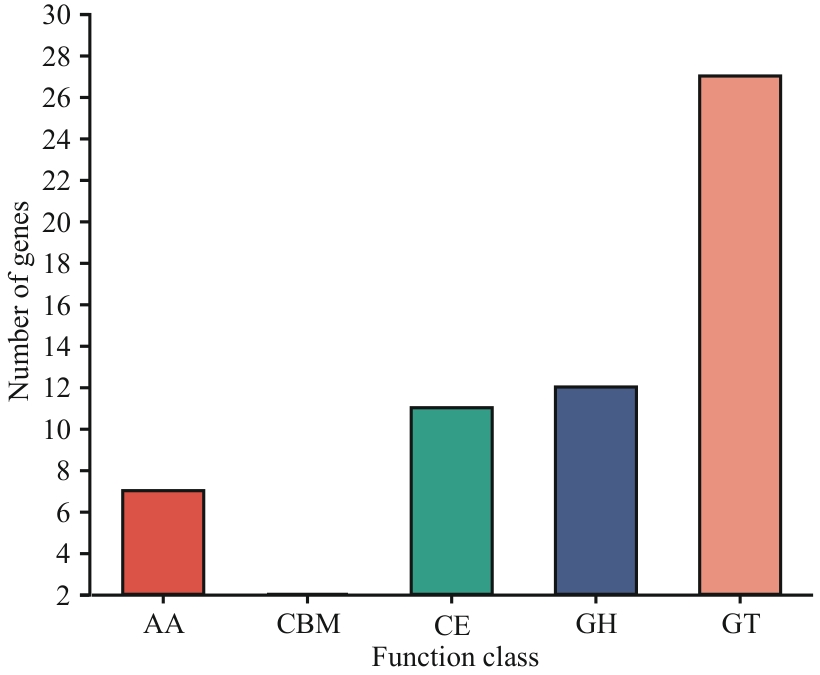
Fig. 11 CAZy analysis of strain FJAT-25102AA: Auxiliary activities. CBM: Carbohydrate-binding modules. CE: Carbohydrat esterases. GH: Glycoside hydrolases. GT: Glycosyl transferases
基因簇编号 Cluster ID | 基因簇类型 Type | 起始位置 Start | 终止位置 End | 已知基因簇 Similar cluster | 相似度 Similarity/% | 基因数量 Gene No. |
|---|---|---|---|---|---|---|
| Cluster1 | Ectoine | 257 860 | 268 232 | - | - | 9 |
| Cluster2 | Siderophore | 1 189 625 | 1 201 461 | - | - | 13 |
| Cluster3 | Betalactone | 1 587 487 | 1 614 450 | 五酮安莎霉素 Microansamycin | 7 | 22 |
| Cluster4 | NRPS-like | 1 653 516 | 1 696 027 | Stenothricin | 31 | 35 |
| Cluster5 | Terpene | 2 371 418 | 2 392 315 | 类胡萝卜素 Carotenoid | 50 | 21 |
Table 4 Predicted results of gene clusters of secondary metabolite of strain FJAT-25102
基因簇编号 Cluster ID | 基因簇类型 Type | 起始位置 Start | 终止位置 End | 已知基因簇 Similar cluster | 相似度 Similarity/% | 基因数量 Gene No. |
|---|---|---|---|---|---|---|
| Cluster1 | Ectoine | 257 860 | 268 232 | - | - | 9 |
| Cluster2 | Siderophore | 1 189 625 | 1 201 461 | - | - | 13 |
| Cluster3 | Betalactone | 1 587 487 | 1 614 450 | 五酮安莎霉素 Microansamycin | 7 | 22 |
| Cluster4 | NRPS-like | 1 653 516 | 1 696 027 | Stenothricin | 31 | 35 |
| Cluster5 | Terpene | 2 371 418 | 2 392 315 | 类胡萝卜素 Carotenoid | 50 | 21 |
基因编号 Gene No. | 基因名称 Gene name | 酶定义 Definition of enzymes | 家族 Family |
|---|---|---|---|
| gene0035 | - | Acetyl xylan esterase (EC 3.1.1.72) | CE7 |
| gene0587 | metX | Acetyl xylan esterase (EC 3.1.1.72) | CE1 |
| gene0588 | - | Acetyl xylan esterase (EC 3.1.1.72) | CE3 |
| gene0589 | - | Acetyl xylan esterase (EC 3.1.1.72) | CE1 |
| gene1502 | yvaK | Acetyl xylan esterase (EC 3.1.1.72) | CE1 |
| gene0137 | malZ | α-glucosidase (EC:3.2.1.20) | GH13 |
| gene0656 | - | Endoglucanase (EC 3.2.1.4) | GH74 |
Table 5 Cellulase-related enzyme families in the strain FJAT-25102 genome
基因编号 Gene No. | 基因名称 Gene name | 酶定义 Definition of enzymes | 家族 Family |
|---|---|---|---|
| gene0035 | - | Acetyl xylan esterase (EC 3.1.1.72) | CE7 |
| gene0587 | metX | Acetyl xylan esterase (EC 3.1.1.72) | CE1 |
| gene0588 | - | Acetyl xylan esterase (EC 3.1.1.72) | CE3 |
| gene0589 | - | Acetyl xylan esterase (EC 3.1.1.72) | CE1 |
| gene1502 | yvaK | Acetyl xylan esterase (EC 3.1.1.72) | CE1 |
| gene0137 | malZ | α-glucosidase (EC:3.2.1.20) | GH13 |
| gene0656 | - | Endoglucanase (EC 3.2.1.4) | GH74 |
| 1 | Ghizzi LG, Del Valle TA, de Castro Zilio EM, et al. Influence of different growing stages of whole-plant soybeans on their nutrient content and silage quality for cattle [J]. Arch Anim Nutr, 2023, 77(6): 437-451. |
| 2 | Zhang XB, He XT, Chen JR, et al. Whole-genome analysis of termite-derived Bacillus velezensis BV-10 and its application in king grass silage [J]. Microorganisms, 2023, 11(11): 2697. |
| 3 | Steiner E, Margesin R. Production and partial characterization of a crude cold-active cellulase (CMCase) from Bacillus mycoides AR20-61 isolated from an Alpine forest site [J]. Ann Microbiol, 2020, 70(1): 67. |
| 4 | Harindintwali JD, Wang F, Yang WH, et al. Harnessing the power of cellulolytic nitrogen-fixing bacteria for biovalorization of lignocellulosic biomass [J]. Ind Crops Prod, 2022, 186: 115235. |
| 5 | 强震宇, 朱林, 朱媛媛, 等. 一株兼具秸秆腐解能力玉米促生菌的筛选、鉴定及发酵优化 [J]. 微生物学通报, 2023, 50(2): 526-540. |
| Qiang ZY, Zhu L, Zhu YY, et al. Isolation and identification of a maize growth-promoting bacterial strain with straw-decomposing capacity and optimization of fermentation conditions [J]. Microbiol China, 2023, 50(2): 526-540. | |
| 6 | 黄悦姝, 邓超, 张馨月, 等. 贝莱斯芽孢杆菌D103产纤维素酶优化及途径分析 [J]. 生物技术, 2023, 33(5): 632-640. |
| Huang YS, Deng C, Zhang XY, et al. Optimization and pathway analysis of cellulase production by Bacillus velezensis D103 [J]. Biotechnology, 2023, 33(5): 632-640. | |
| 7 | 孟建宇, 李蘅, 樊兆阳, 等. 低温纤维素降解菌的分离与鉴定 [J]. 应用与环境生物学报, 2014, 20(1): 152-156. |
| Meng JY, Li H, Fan ZY, et al. Isolation and identification of cellulose-degrading bacteria under low temperature [J]. Chin J Appl Environ Biol, 2014, 20(1): 152-156. | |
| 8 | 黄婉秋, 石冬冬, 蔡红英, 等. 白星花金龟幼虫肠道中纤维素降解菌的筛选及其全基因组分析 [J]. 中国农业科技导报, 2021, 23(6): 51-58. |
| Huang WQ, Shi DD, Cai HY, et al. Identification and genome analysis of a cellulose degrading strain from the intestinal tract of Protaetia brevitarsis larva [J]. J Agric Sci Technol, 2021, 23(6): 51-58. | |
| 9 | 李国强, 罗巧玉, 杨梅娇, 等. 松墨天牛肠道BS16基因组及其纤维素酶活特性分析 [J]. 西南林业大学学报: 自然科学, 2023, 43(4): 100-108. |
| Li GQ, Luo QY, Yang MJ, et al. Genome analysis of Monochamus alternatus gut BS16 and characterization of its cellulase activity [J]. J Southwest For Univ Nat Sci, 2023, 43(4): 100-108. | |
| 10 | Raza FA, Faisal M. Growth promotion of maize by desiccation tolerant Micrococcus luteus-chp37 isolated from Cholistan desert, Pakistan [J]. Aust J Crop Sci, 2013, 7(11): 1693-1698. |
| 11 | Jampasri K, Pokethitiyook P, Kruatrachue M, et al. Phytoremediation of fuel oil and lead co-contaminated soil by Chromolaena odorata in association with Micrococcus luteus [J]. Int J Phytoremediation, 2016, 18(10): 994-1001. |
| 12 | Jampasri K, Pokethitiyook P, Poolpak T, et al. Bacteria-assisted phytoremediation of fuel oil and lead co-contaminated soil in the salt-stressed condition by Chromolaena odorata and Micrococcus luteus [J]. Int J Phytoremediation, 2020, 22(3): 322-333. |
| 13 | 李云琪, 王宇辉, 李小锦, 等. 藤黄微球菌Rpf因子对土壤细菌可培养物种分离效果的影响 [J]. 河北大学学报: 自然科学版, 2019, 39(1): 63-68. |
| Li YQ, Wang YH, Li XJ, et al. Effects of Rpf factor of Micrococcus luteus on the isolation of soil culturable species [J]. J Hebei Univ Nat Sci Ed, 2019, 39(1): 63-68. | |
| 14 | 陈曦, 陈红, 刘懋, 等. 藤黄微球菌(Micrococcus luteus)KDF1胞外蛋白酶酶学性质与水解大豆蛋白产物研究 [J]. 食品与发酵工业, 2013, 39(9): 11-17. |
| Chen X, Chen H, Liu M, et al. The extracellular protease from Micrococcus luteus KDF1: properties and characterization of its hydrolysates from soy protein [J]. Food Ferment Ind, 2013, 39(9): 11-17. | |
| 15 | 宋时丽, 吴昊, 黄鹏伟, 等. 秸秆还田土壤改良培肥基质和复合菌剂配施对土壤生态的影响 [J]. 生态学报, 2021, 41(11): 4562-4576. |
| Song SL, Wu H, Huang PW, et al. Effects of total straw incorporation combined with soil modified fertilizer substrate and compound microbial agent on soil ecology and wheat yield [J]. Acta Ecol Sin, 2021, 41(11): 4562-4576. | |
| 16 | 王永伦, 余克非, 郑展望. 1株耐高温纤维素降解菌发酵条件优化与秸秆降解应用 [J]. 江苏农业科学, 2023, 51(19): 229-236, 244. |
| Wang YL, Yu KF, Zheng ZW. Optimization of fermentation conditions of a cellulose degrading strain with high temperature resistance and its application in straw degradation [J]. Jiangsu Agric Sci, 2023, 51(19): 229-236, 244. | |
| 17 | 魏越波, 郭晓军, 孙悦龙, 等. 发酵用高温放线菌的筛选及性质研究 [J]. 饲料工业, 2020, 41(21): 47-52. |
| Wei YB, Guo XJ, Sun YL, et al. Screening and properties of high-temperature actinomycetes for fermentation [J]. Feed Ind, 2020, 41(21): 47-52. | |
| 18 | Lichev A, Angelov A, Cucurull I, et al. Amino acids as nutritional factors and (p)ppGpp as an alarmone of the stringent response regulate natural transformation in Micrococcus luteus [J]. Sci Rep, 2019, 9: 11030. |
| 19 | Sher S, Rehman A, Hansen LH, et al. Complete genome sequences of highly arsenite-resistant bacteria Brevibacterium sp. strain CS2 and Micrococcus luteus AS2 [J]. Microbiol Resour Announc, 2019, 8(31): e00531-19. |
| 20 | 孙敏, 陈天宇, 冯红. 藤黄微球菌V017基因组测序及对辐照的转录组响应 [J]. 微生物学通报, 2021, 48(5): 1648-1661. |
| Sun M, Chen TY, Feng H. Complete genome of Micrococcus luteus V017 and its transcriptomic response to gamma ray radiation [J]. Microbiol China, 2021, 48(5): 1648-1661. | |
| 21 | Kutmutia SK, Drautz-Moses DI, Uchida A, et al. Complete genome sequence of Micrococcus luteus strain SGAir0127, isolated from indoor air samples from Singapore [J]. Microbiol Resour Announc, 2019, 8(41): e00646-19. |
| 22 | Martínez FL, Anton BP, DasSarma P, et al. Complete genome sequence and methylome analysis of Micrococcus luteus SA211, a halophilic, lithium-tolerant actinobacterium from Argentina [J]. Microbiol Resour Announc, 2019, 8(4): e01557-18. |
| 23 | Wang J, Lu C, Tang Y, et al. Microansamycins J and K from Micromonospora sp. HK160111mas13OE [J]. Nat Prod Res, 2024, 38(21): 3854-3858. |
| 24 | Liu S, Gao YW, Quan L, et al. Improvement of lignocellulolytic enzyme production mediated by calcium signaling in Bacillus subtilis Z2 under graphene oxide stress [J]. Appl Environ Microbiol, 2022, 88(19): e0096022. |
| 25 | 饶紫环, 谢志雄. 一株Olivibacter jilunii纤维素降解菌株的分离鉴定与降解能力分析 [J]. 生物技术通报, 2023, 39(8): 283-290. |
| Rao ZH, Xie ZX. Isolation and identification of a cellulose-degrading strain of Olivibacter jilunii and analysis of its degradability [J]. Biotechnol Bull, 2023, 39(8): 283-290. | |
| 26 | 张超, 李晓意, 杨吉, 等. 1株耐酸秸秆纤维素降解菌株的筛选、鉴定及其内切葡聚糖酶的异源表达研究 [J]. 饲料研究, 2023, 46(2): 89-93. |
| Zhang C, Li XY, Yang J, et al. Study on screening and identification of an acid-tolerant straw cellulose-degrading bacteria and heterologous expression of endoglucanase [J]. Feed Res, 2023, 46(2): 89-93. | |
| 27 | Zafar A, Aftab MN, Din ZU, et al. Cloning, expression, and purification of xylanase gene from Bacillus licheniformis for use in saccharification of plant biomass [J]. Appl Biochem Biotechnol, 2016, 178(2): 294-311. |
| 28 | 王靖宇, 刘玉春, 韩伟, 等. 玉米皮纤维发酵裂褶菌的产酶分析及木聚糖酶基因克隆、表达和酶学性质测定 [J]. 食品与发酵工业, 2018, 44(5): 46-51. |
| Wang JY, Liu YC, Han W, et al. Analysis on enzyme production of Schizophyllum commune fermented with corn bran fiber and cloning, expression and characterization of xylanase gene [J]. Food Ferment Ind, 2018, 44(5): 46-51. | |
| 29 | Yang R, Li JC, Teng C, et al. Cloning, overexpression and characterization of a xylanase gene from a novel Streptomyces rameus L2001 in Pichia pastoris [J]. J Mol Catal B Enzym, 2016, 131: 85-93. |
| 30 | He LW, Zhou W, Wang C, et al. Effect of cellulase and Lactobacillus casei on ensiling characteristics, chemical composition, antioxidant activity, and digestibility of mulberry leaf silage [J]. J Dairy Sci, 2019, 102(11): 9919-9931. |
| [1] | SONG Shu-yi, JIANG Kai-xiu, LIU Huan-yan, HUANG Ya-cheng, LIU Lin-ya. Identification of the TCP Gene Family in Actinidia chinensis var. Hongyang and Their Expression Analysis in Fruit [J]. Biotechnology Bulletin, 2025, 41(3): 190-201. |
| [2] | LI Bin, SU Xiang-ping, LIU Chang, WANG Yu-bing, ZHANG Yong-hong, ZHOU Chao, XU Qing. Chloroplast Genome Characteristics and Phylogenetic Analysis of Scrophulariaceae [J]. Biotechnology Bulletin, 2025, 41(3): 240-254. |
| [3] | SONG Ying-pei, WANG Can, ZHOU Hui-wen, KONG Ke-ke, XU Meng-ge, WANG Rui-kai. Analysis of Soybean Pod Dehiscence Habit Based on Whole Genome Association Analysis and Genetic Diversity [J]. Biotechnology Bulletin, 2025, 41(2): 97-106. |
| [4] | HE Cai-lin, LU Jing, GUO Hui-hui, LI Xiao-an, WU Qi. Genome-wide Identification and Expression Analysis of the MADS-box Gene Family in Quinoa [J]. Biotechnology Bulletin, 2025, 41(1): 157-172. |
| [5] | ZHANG Ting, WAN Yu-xin, XU Wei-hui, WANG Zhi-gang, CHEN Wen-jing, HU Yun-long. Growth-promoting Effects of a Rhizosphere Growth-promoting Bacterium Leclercia adecarboxylata LN01 in Maize Plants and Its Whole-genome Analysis [J]. Biotechnology Bulletin, 2025, 41(1): 263-275. |
| [6] | JIANG Hong-yan, CHEN Shi-chun, LIAO Shu-ran, CHEN Ting-xu, WANG Xiao-qing. The Complete Sequences and Phylogenetic Analysis of Mitochondrial Genomes in Eocanthecona concinna and Picromerus lewisi [J]. Biotechnology Bulletin, 2025, 41(1): 312-323. |
| [7] | JIN Su-kui, GUO Qian-qian, LIU Qiao-quan, GAO Ji-ping. A Simplified Method for Extracting Genomic DNA from Rice Leaves [J]. Biotechnology Bulletin, 2025, 41(1): 74-84. |
| [8] | SONG Qian-na, DUAN Yong-hong, FENG Rui-yun. Establishment of CRISPR/Cas9-mediated Highly Efficient Gene Editing System in Microtubers of Potatoes [J]. Biotechnology Bulletin, 2024, 40(9): 33-41. |
| [9] | CUI Yuan-yuan, WANG Zhao-yi, BAI Shuang-yu, REN Yu-zhao, DOU Fei-fei, LIU Cai-xia, LIU Feng-lou, WANG Zhang-jun, LI Qing-feng. Genome-wide Identification of Non-specific Phospholipase C Gene Family in Hordeum vulgare L. and Stress Expression Analysis at Seedling Stage [J]. Biotechnology Bulletin, 2024, 40(8): 74-82. |
| [10] | SUN Zhi-yong, DU Huai-dong, LIU Yang, MA Jia-xin, YU Xue-ran, MA Wei, YAO Xin-jie, WANG Min, LI Pei-fu. Genome-wide Association Analysis of γ-aminobutyric Acid in Rice Grains [J]. Biotechnology Bulletin, 2024, 40(8): 53-62. |
| [11] | ZANG Wen-rui, MA Ming, CHE Gen, HASI Agula. Genome-wide Identification and Expression Pattern Analysis of BZR Transcription Factor Gene Family of Melon [J]. Biotechnology Bulletin, 2024, 40(7): 163-171. |
| [12] | ZHOU Jiang-hong, XIA Fei, ZHONG Li, QIU Lan-fen, LI Guang, LIU Qian, ZHANG Guo-feng, SHAO Jin-li, LI Na, CHE Shao-chen. Whole Genome Sequencing and Comparative Genomic Analysis of Antagonistic Bacterium CCBC3-3-1 against Verticillium dahlia [J]. Biotechnology Bulletin, 2024, 40(7): 235-246. |
| [13] | TIAN Tong-tong, GE Jia-zhen, GAO Peng-cheng, LI Xue-rui, SONG Guo-dong, ZHENG Fu-ying, CHU Yue-feng. Whole Genome Sequencing and Bioinformatics Analysis of Mycoplasma ovipneumoniae GH3-3 Strain [J]. Biotechnology Bulletin, 2024, 40(7): 323-334. |
| [14] | ALIYA Waili, CHEN Yong-kun, KELAREMU Kelimujiang, WANG Bao-qing, CHEN Ling-na. Phylogenetic Evolution and Expression Analysis of SPL Gene Family in Juglans regia [J]. Biotechnology Bulletin, 2024, 40(6): 180-189. |
| [15] | KONG Xiao-ping, CHEN Li-wen, LIU Si-si, YAN Xiang-ping. Genome-wide Association Study of Bolting Related Traits in Carrot [J]. Biotechnology Bulletin, 2024, 40(5): 120-130. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||