








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (10): 288-295.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0455
Previous Articles Next Articles
LONG Jia-jia1,2,3( ), LIU Wei-wei1,2,3, FAN Xin-hao2,3,4, LI Wang-chang1, YANG Xiao-gan1, TANG Zhong-lin1,2,3,4(
), LIU Wei-wei1,2,3, FAN Xin-hao2,3,4, LI Wang-chang1, YANG Xiao-gan1, TANG Zhong-lin1,2,3,4( )
)
Received:2024-05-15
Online:2024-10-26
Published:2024-11-20
Contact:
TANG Zhong-lin
E-mail:2118301022@st.gxu.edu.cn;tangzhonglin@caas.cn
LONG Jia-jia, LIU Wei-wei, FAN Xin-hao, LI Wang-chang, YANG Xiao-gan, TANG Zhong-lin. Study on Constructing an RNA Editing Map of Pig Based on Large-scale RNA-seq Data[J]. Biotechnology Bulletin, 2024, 40(10): 288-295.
| 组织 Tissue | RNA-seq数据量 Number of RNA-seq data | 过滤阈值 Filtering threshold |
|---|---|---|
| 骨骼肌Skeletal muscle | 1427 | 16 |
| 肝Liver | 367 | 6 |
| 大肠Large intestine | 74 | 6 |
| 大脑Brain | 215 | 9 |
| 肺Lung | 118 | 11 |
| 小肠Small intestine | 239 | 11 |
| 卵巢Ovary | 99 | 5 |
| 输卵管Oviduct | 22 | 5 |
| 脂肪Adipose | 332 | 16 |
| 心Heart | 89 | 5 |
| 睾丸Testis | 133 | 7 |
| 子宫Uterus | 150 | 7 |
| 淋巴Lymph | 32 | 5 |
| 垂体Pituitary | 52 | 8 |
| 脾Spleen | 78 | 5 |
| 肾Kindy | 34 | 5 |
Table 1 Filtering criteria for RNA editing sites of all tissues
| 组织 Tissue | RNA-seq数据量 Number of RNA-seq data | 过滤阈值 Filtering threshold |
|---|---|---|
| 骨骼肌Skeletal muscle | 1427 | 16 |
| 肝Liver | 367 | 6 |
| 大肠Large intestine | 74 | 6 |
| 大脑Brain | 215 | 9 |
| 肺Lung | 118 | 11 |
| 小肠Small intestine | 239 | 11 |
| 卵巢Ovary | 99 | 5 |
| 输卵管Oviduct | 22 | 5 |
| 脂肪Adipose | 332 | 16 |
| 心Heart | 89 | 5 |
| 睾丸Testis | 133 | 7 |
| 子宫Uterus | 150 | 7 |
| 淋巴Lymph | 32 | 5 |
| 垂体Pituitary | 52 | 8 |
| 脾Spleen | 78 | 5 |
| 肾Kindy | 34 | 5 |

Fig. 1 Identification result and validation of RNA editing sites A:PCA analysis plot of RNA editing sites across all tissues. B:Distribution of all RNA editing site types and repeat regions. C:Venn diagram of sites from this study and other studies. D:Validation of RNA editing sites by sanger, the blue arrows indicates the locations of RNA editing sites
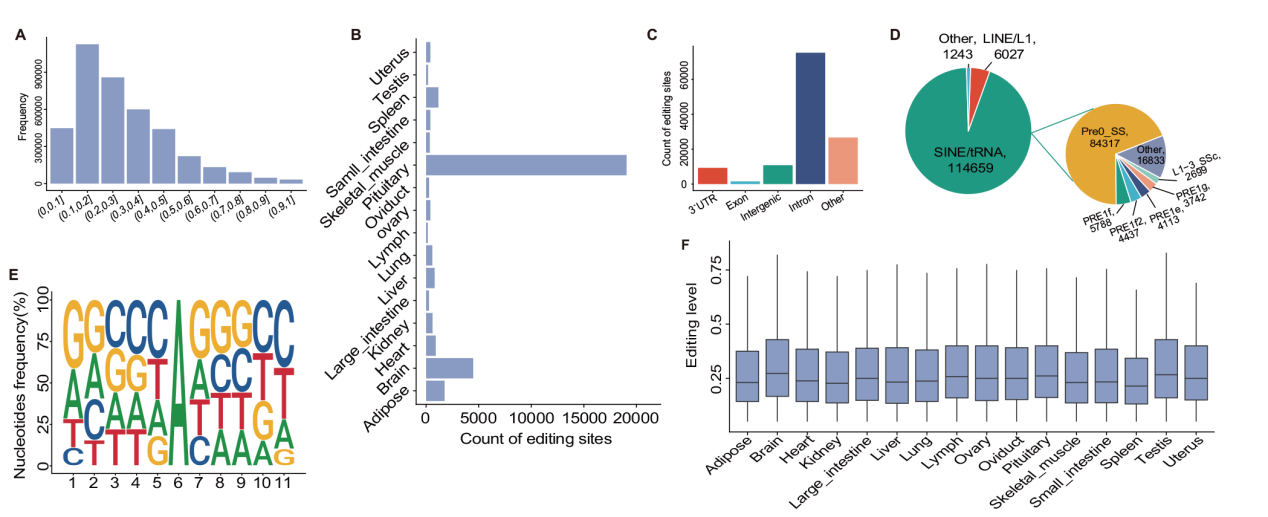
Fig. 2 Characteristics of A-to-I RNA editing sites across all tissues A: Histogram of overall editing levels of A-to-I RNA editing sites. B:Average number of sites per sample in different tissues. C:Histogram of the genomic distribution of A-to-I RNA editing sites. D:Distribution of A-to-I RNA editing sites in repeat regions. E:Upstream and downstream 5 bp base preference analysis of A-to-I RNA editing sites. F:Editing levels of sites in different tissues
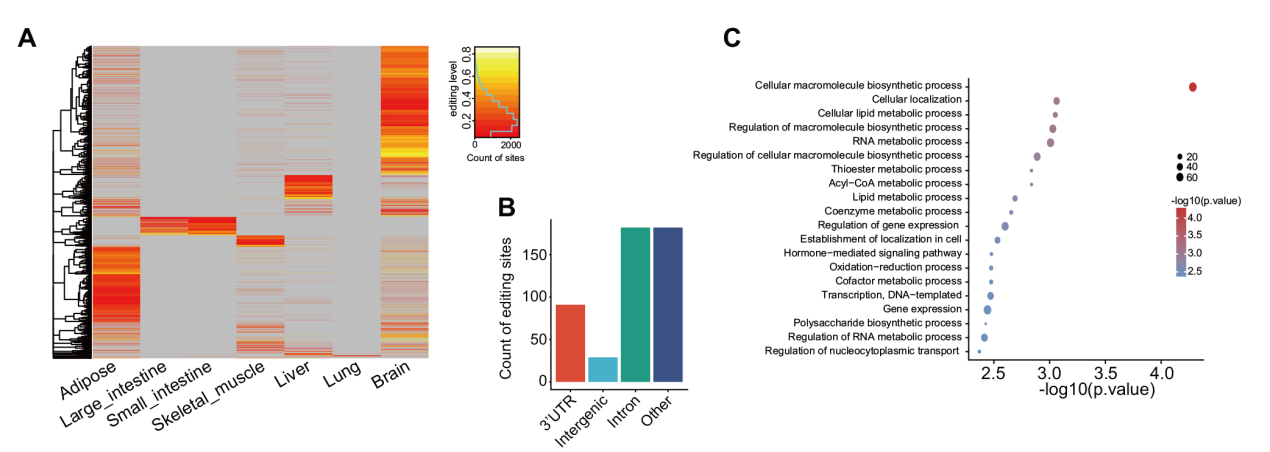
Fig. 4 Specific sites of adipose and other tissue A:Heatmap of tissue-specific sites in seven tissues. B:Histogram of the genomic distribution of adipose tissue-specific sites. C:Bar chart of GO enrichment of host genes for adipose tissue-specific sites
| [1] |
Gabay O, Shoshan Y, Kopel E, et al. Landscape of adenosine-to-inosine RNA recoding across human tissues[J]. Nat Commun, 2022, 13(1): 1184.
doi: 10.1038/s41467-022-28841-4 pmid: 35246538 |
| [2] |
Noda Y, Okada S, Suzuki T. Regulation of A-to-I RNA editing and stop codon recoding to control selenoprotein expression during skeletal myogenesis[J]. Nat Commun, 2022, 13(1): 2503.
doi: 10.1038/s41467-022-30181-2 pmid: 35523818 |
| [3] |
Teoh PJ, Koh MY, Chng WJ. ADARs, RNA editing and more in hematological malignancies[J]. Leukemia, 2021, 35(2): 346-359.
doi: 10.1038/s41375-020-01076-2 pmid: 33139858 |
| [4] |
Jimeno S, Prados-Carvajal R, Fernández-Ávila MJ, et al. ADAR-mediated RNA editing of DNA: RNA hybrids is required for DNA double strand break repair[J]. Nat Commun, 2021, 12(1): 5512.
doi: 10.1038/s41467-021-25790-2 pmid: 34535666 |
| [5] |
Shiromoto Y, Sakurai M, Minakuchi M, et al. ADAR1 RNA editing enzyme regulates R-loop formation and genome stability at telomeres in cancer cells[J]. Nat Commun, 2021, 12(1): 1654.
doi: 10.1038/s41467-021-21921-x pmid: 33712600 |
| [6] | Heraud-Farlow JE, Walkley CR. What do editors do? Understanding the physiological functions of A-to-I RNA editing by adenosine deaminase acting on RNAs[J]. Open Biol, 2020, 10(7): 200085. |
| [7] |
Ma YY, Dammer EB, Felsky D, et al. Atlas of RNA editing events affecting protein expression in aged and Alzheimer's disease human brain tissue[J]. Nat Commun, 2021, 12(1): 7035.
doi: 10.1038/s41467-021-27204-9 pmid: 34857756 |
| [8] | Lebrigand K, Bergenstråhle J, Thrane K, et al. The spatial landscape of gene expression isoforms in tissue sections[J]. Nucleic Acids Res, 2023, 51(8): e47. |
| [9] | Wu D, Zang YY, Shi YY, et al. Distant coupling between RNA editing and alternative splicing of the osmosensitive cation channel Tmem63b[J]. J Biol Chem, 2020, 295(52): 18199-18212. |
| [10] | Voss G, Cassidy JR, Ceder Y. Functional consequences of A-to-I editing of miR-379 in prostate cancer cells[J]. Sci Rep, 2023, 13(1): 16602. |
| [11] |
Shen HQ, An O, Ren X, et al. ADARs act as potent regulators of circular transcriptome in cancer[J]. Nat Commun, 2022, 13(1): 1508.
doi: 10.1038/s41467-022-29138-2 pmid: 35314703 |
| [12] | Xu BY, Qin WX, Chen YW, et al. Multi-omics analysis reveals gut microbiota-ovary axis contributed to the follicular development difference between Meishan and Landrace × Yorkshire sows[J]. J Anim Sci Biotechnol, 2023, 14(1): 68. |
| [13] |
Zhou YY, Li JQ, Huang F, et al. Characterization of the pig lower respiratory tract antibiotic resistome[J]. Nat Commun, 2023, 14(1): 4868.
doi: 10.1038/s41467-023-40587-1 pmid: 37573429 |
| [14] |
Ling ZQ, Li J, Jiang T, et al. Omics-based construction of regulatory variants can be applied to help decipher pig liver-related traits[J]. Commun Biol, 2024, 7(1): 381.
doi: 10.1038/s42003-024-06050-7 pmid: 38553586 |
| [15] | Adetula AA, Fan XH, Zhang YS, et al. Landscape of tissue-specific RNA Editome provides insight into co-regulated and altered gene expression in pigs(Sus-scrofa)[J]. RNA Biol, 2021, 18(sup1): 439-450. |
| [16] | Paukszto L, Mikolajczyk A, Jastrzebski JP, et al. Transcriptome, spliceosome and editome expression patterns of the porcine endometrium in response to a single subclinical dose of Salmonella enteritidis lipopolysaccharide[J]. Int J Mol Sci, 2020, 21(12): 4217. |
| [17] | Yang L, Huang L, Mu YL, et al. Characterization of A-to-I editing in pigs under a long-term high-energy diet[J]. Int J Mol Sci, 2023, 24(9): 7921. |
| [18] |
Li TT, Li Q, Li H, et al. Pig-specific RNA editing during early embryo development revealed by genome-wide comparisons[J]. FEBS Open Bio, 2020, 10(7): 1389-1402.
doi: 10.1002/2211-5463.12900 pmid: 32433824 |
| [19] | Yang YL, Zhu M, Fan XH, et al. Developmental atlas of the RNA editome in Sus scrofa skeletal muscle[J]. DNA Res, 2019, 26(3): 261-272. |
| [20] | Wang ZS, Feng XK, Tang ZL, et al. Genome-wide investigation and functional analysis of Sus scrofa RNA editing sites across eleven tissues[J]. Genes, 2019, 10(5): 327. |
| [21] | Tan MH, Consortium G, Li Q, et al. Dynamic landscape and regulation of RNA editing in mammals[J]. Nature, 2017, 550(7675): 249-254. |
| [22] | Zhang YB, Zhang LC, Yue JW, et al. Genome-wide identification of RNA editing in seven porcine tissues by matched DNA and RNA high-throughput sequencing[J]. J Anim Sci Biotechnol, 2019, 10: 24. |
| [23] |
Sherman BT, Hao M, Qiu J, et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists(2021 update)[J]. Nucleic Acids Res, 2022, 50(W1): W216-W221.
doi: 10.1093/nar/gkac194 pmid: 35325185 |
| [24] | Larsen K, Heide-Jørgensen MP. Conservation of A-to-I RNA editing in bowhead whale and pig[J]. PLoS One, 2021, 16(12): e0260081. |
| [25] | Zhou R, Yao WY, Xie CD, et al. Developmental stage-specific A-to-I editing pattern in the postnatal pineal gland of pigs(Sus scrofa)[J]. J Anim Sci Biotechnol, 2020, 11: 90. |
| [26] |
Nguyen TA, Heng JWJ, Kaewsapsak P, et al. Direct identification of A-to-I editing sites with nanopore native RNA sequencing[J]. Nat Methods, 2022, 19(7): 833-844.
doi: 10.1038/s41592-022-01513-3 pmid: 35697834 |
| [27] | Wu XY, Ayalew W, Chu M, et al. Characterization of RNA editome in the mammary gland of yaks during the lactation and dry periods[J]. Animals, 2022, 12(2): 207. |
| [28] |
Booth BJ, Nourreddine S, Katrekar D, et al. RNA editing: expanding the potential of RNA therapeutics[J]. Mol Ther, 2023, 31(6): 1533-1549.
doi: 10.1016/j.ymthe.2023.01.005 pmid: 36620962 |
| [29] |
Zhang F, Lu YL, Yan SJ, et al. SPRINT: an SNP-free toolkit for identifying RNA editing sites[J]. Bioinformatics, 2017, 33(22): 3538-3548.
doi: 10.1093/bioinformatics/btx473 pmid: 29036410 |
| [30] |
Zambrano-Mila MS, Witzenberger M, Rosenwasser Z, et al. Dissecting the basis for differential substrate specificity of ADAR1 and ADAR2[J]. Nat Commun, 2023, 14(1): 8212.
doi: 10.1038/s41467-023-43633-0 pmid: 38081817 |
| [31] |
Huang JR, Lin L, Dong ZY, et al. A porcine brain-wide RNA editing landscape[J]. Commun Biol, 2021, 4(1): 717.
doi: 10.1038/s42003-021-02238-3 pmid: 34112917 |
| [32] |
Yao L, Wang HM, Song YY, et al. Large-scale prediction of ADAR-mediated effective human A-to-I RNA editing[J]. Brief Bioinform, 2019, 20(1): 102-109.
doi: 10.1093/bib/bbx092 pmid: 28968662 |
| [33] |
Al-Roub A, Akhter N, Al-Rashed F, et al. TNFα induces matrix metalloproteinase-9 expression in monocytic cells through ACSL1/JNK/ERK/NF-kB signaling pathways[J]. Sci Rep, 2023, 13(1): 14351.
doi: 10.1038/s41598-023-41514-6 pmid: 37658104 |
| [34] | Zhang XY, Gao Y, Liu ZZ, et al. Salicylate sodium suppresses monocyte chemoattractant protein-1 production by directly inhibiting phosphodiesterase 3B in TNF-α-stimulated adipocytes[J]. Int J Mol Sci, 2022, 24(1): 320. |
| [35] | Hosooka T, Hosokawa Y, Matsugi K, et al. The PDK1-FoxO1 signaling in adipocytes controls systemic insulin sensitivity through the 5-lipoxygenase-leukotriene B4 axis[J]. Proc Natl Acad Sci USA, 2020, 117(21): 11674-11684. |
| [36] | Liebscher G, Vujic N, Schreiber R, et al. The lysosomal LAMTOR/Ragulator complex is essential for nutrient homeostasis in brown adipose tissue[J]. Mol Metab, 2023, 71: 101705. |
| [1] | ALIYA Waili, CHEN Yong-kun, KELAREMU Kelimujiang, WANG Bao-qing, CHEN Ling-na. Phylogenetic Evolution and Expression Analysis of SPL Gene Family in Juglans regia [J]. Biotechnology Bulletin, 2024, 40(6): 180-189. |
| [2] | CHANG Xue-rui, WANG Tian-tian, WANG Jing. Identification and Analysis of E2 Gene Family in Pepper(Capsicum annuum L.) [J]. Biotechnology Bulletin, 2024, 40(6): 238-250. |
| [3] | WANG Di ZHANG Xiao-yu SONG Yu-xin ZHENG Dong-ran TIAN Jing LI Yu-hua WANG Yu WU Hao. Advances in the Molecular Mechanisms of Plant Tissue Culture and Regeneration Regulated by Totipotency-related Transcription Factors [J]. Biotechnology Bulletin, 2024, 40(6): 23-33. |
| [4] | LIU Bao-cai, ZHANG Wu-jun, ZHAO Yun-qing, HUANG Ying-zhen, CHEN Jing-ying. Identification and Tissue Expression of Four Transcription Factors in Pholidota chinensis [J]. Biotechnology Bulletin, 2024, 40(5): 141-152. |
| [5] | LU Yu-dan, LIU Xiao-chi, FENG Xin, CHEN Gui-xin, CHEN Yi-ting. Identification of the Kiwifruit BBX Gene Family and Analysis of Their Transcriptional Characteristics [J]. Biotechnology Bulletin, 2024, 40(2): 172-182. |
| [6] | LI Can, JIANG Xiang-ning, GAI Ying. Cloning of the LkF3H2 Gene in Larix kaempferi and Its Function in Regulating Flavonoid Metabolism [J]. Biotechnology Bulletin, 2024, 40(2): 245-252. |
| [7] | WANG Feng-ting, ZHAO Fu-shun, QIAO Kai-bin, XU Xun, LIU Jin-liang. Progress on the Molecular Mechanism of Scion-rootstock Interactions in Vegetable Grafting [J]. Biotechnology Bulletin, 2024, 40(10): 149-159. |
| [8] | YANG Shuai-peng, QU Zi-xiao, ZHU Xiang-xing, TANG Dong-sheng. Optimization of DNA Base Editing Technology and Its Application in Pig Genetic Modification [J]. Biotechnology Bulletin, 2024, 40(1): 127-144. |
| [9] | ZHANG Jin-wei, WU Yuan-xia, SUN Jing, LI Xiao-kai, LU Lu, LI Zhou-quan, GE Liang-peng. Effects of Commensal Microbiota on Intestinal Development, Metabolism, and Mitochondrial Function in Piglets [J]. Biotechnology Bulletin, 2024, 40(1): 332-343. |
| [10] | ZHANG Dao-lei, GAN Yu-jun, LE Liang, PU Li. Epigenetic Regulation of Yield-related Traits in Maize and Epibreeding [J]. Biotechnology Bulletin, 2023, 39(8): 31-42. |
| [11] | LI Ying, YUE Xiang-hua. Application of DNA Methylation in Interpreting Natural Variation in Moso Bamboo [J]. Biotechnology Bulletin, 2023, 39(7): 48-55. |
| [12] | WANG Bing, ZHAO Hui-na, YU Jing, YU Shi-zhou, LEI Bo. Research Progress in the Regulation of Plant Branch Development [J]. Biotechnology Bulletin, 2023, 39(5): 14-22. |
| [13] | LI Yi-jun, WU Chen-chen, LI Rui, WANG Zhe, HE Shan-wen, WEI Shan-jun, ZHANG Xiao-xia. Exploring Cultivation Approaches for New Endophytic Bacterial Resource in Oryza sativa [J]. Biotechnology Bulletin, 2023, 39(4): 201-211. |
| [14] | WEI Ming WANG Xin-yu WU Guo-qiang ZHAO Meng. The Role of NAD-dependent Deacetylase SRT in Plant Epigenetic Inheritance Regulation [J]. Biotechnology Bulletin, 2023, 39(4): 59-70. |
| [15] | CHEN Chu-wen, LI Jie, ZHAO Rui-peng, LIU Yuan, WU Jin-bo, LI Zhi-xiong. Cloning, Tissue Expression Profile and Function Prediction of GPX3 Gene in Tibetan Chicken [J]. Biotechnology Bulletin, 2023, 39(3): 311-320. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||