








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (4): 306-318.doi: 10.13560/j.cnki.biotech.bull.1985.2023-1048
Previous Articles Next Articles
ZHANG Qing-lan1,2( ), ZHANG Ya-ran2,3(
), ZHANG Ya-ran2,3( ), JU Zhi-hua2,3, WANG Xiu-ge2,3, XIAO Yao2,3, WANG Jin-peng2,3, WEI Xiao-chao2,3, GAO Ya-ping2,3, BAI Fu-heng4, WANG Hong-cheng1(
), JU Zhi-hua2,3, WANG Xiu-ge2,3, XIAO Yao2,3, WANG Jin-peng2,3, WEI Xiao-chao2,3, GAO Ya-ping2,3, BAI Fu-heng4, WANG Hong-cheng1( )
)
Received:2023-11-09
Online:2024-04-26
Published:2024-04-30
Contact:
ZHANG Ya-ran, WANG Hong-cheng
E-mail:1400304672@qq.com;zhang_ya_ran@126.com;wanghc@gznu.edu.cn
ZHANG Qing-lan, ZHANG Ya-ran, JU Zhi-hua, WANG Xiu-ge, XIAO Yao, WANG Jin-peng, WEI Xiao-chao, GAO Ya-ping, BAI Fu-heng, WANG Hong-cheng. Identification and Transcriptional Regulation Analysis of Core Promoter in Bovine TARDBP Gene[J]. Biotechnology Bulletin, 2024, 40(4): 306-318.
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 产物大小 Product length/bp | 退火温度 Annealing temperature/℃ | 用途 Usage |
|---|---|---|---|---|
| TARDBP-F | ATGTCTGAATATATTCGGGTAACCG | 1 245 | 64.4 | CDS区扩增 |
| TARDBP-R | CTACATTCCCCAGCCAGAAGATTTA | |||
| TARDBP-GSP | CCGTGAAGCGAACGAAGCCAAACC | 496 | 68 | 5'RACE第一轮PCR |
| TARDBP -NGSP | AGATTCCCCCAGCCAGCATCGG | 230 | 5'RACE第二轮PCR | |
| pTARDBP-F | GCAAAGGGATAGAGGGAA | 1 954 | 55 | 启动子区扩增 |
| pTARDBP-R | CAGGGCGTCCTTAGTGAG | |||
| pGL3-P1 | CGG GGTACCGCAAAGGGATAGAGGGAA | 1 954 | 55 | 不同缺失片段扩增 |
| pGL3-P2 | CGG GGTACCGTTCAAACTACCGCACAATG | 1 498 | ||
| pGL3-P3 | CGG GGTACCTAGGGTGGCAAAGAGTTGG | 1 179 | ||
| pGL3-P4 | CGG GGTACCGAGAAGGAAATGGCAACCC | 854 | ||
| pGL3-P5 | CGG GGTACCAATGGCTTGGGCTCAACAG | 615 | ||
| pGL3-P6 | CGG GGTACCTATAGCCTTCAAGTTCACTCCC | 288 | ||
| pGL3-R1 | CCC AAGCTTCAGGGCGTCCTTAGTGAG | |||
| TARDBP-q-F | GTTTGGCTTCGTTCGCTTCA | 164 | 60 | RT-qPCR |
| TARDBP-q-R | CCTCTGTACAACGCCCAACA | |||
| β-actin-q-F | CATCGGCAATGAGCGGTTCC | 147 | ||
| β-actin-q-R | ACCGTGTTGGCGTAGAGGTC |
Table 1 Sequence information of bovine TARDBP gene primer
| 引物名称 Primer name | 引物序列 Primer sequence(5'-3') | 产物大小 Product length/bp | 退火温度 Annealing temperature/℃ | 用途 Usage |
|---|---|---|---|---|
| TARDBP-F | ATGTCTGAATATATTCGGGTAACCG | 1 245 | 64.4 | CDS区扩增 |
| TARDBP-R | CTACATTCCCCAGCCAGAAGATTTA | |||
| TARDBP-GSP | CCGTGAAGCGAACGAAGCCAAACC | 496 | 68 | 5'RACE第一轮PCR |
| TARDBP -NGSP | AGATTCCCCCAGCCAGCATCGG | 230 | 5'RACE第二轮PCR | |
| pTARDBP-F | GCAAAGGGATAGAGGGAA | 1 954 | 55 | 启动子区扩增 |
| pTARDBP-R | CAGGGCGTCCTTAGTGAG | |||
| pGL3-P1 | CGG GGTACCGCAAAGGGATAGAGGGAA | 1 954 | 55 | 不同缺失片段扩增 |
| pGL3-P2 | CGG GGTACCGTTCAAACTACCGCACAATG | 1 498 | ||
| pGL3-P3 | CGG GGTACCTAGGGTGGCAAAGAGTTGG | 1 179 | ||
| pGL3-P4 | CGG GGTACCGAGAAGGAAATGGCAACCC | 854 | ||
| pGL3-P5 | CGG GGTACCAATGGCTTGGGCTCAACAG | 615 | ||
| pGL3-P6 | CGG GGTACCTATAGCCTTCAAGTTCACTCCC | 288 | ||
| pGL3-R1 | CCC AAGCTTCAGGGCGTCCTTAGTGAG | |||
| TARDBP-q-F | GTTTGGCTTCGTTCGCTTCA | 164 | 60 | RT-qPCR |
| TARDBP-q-R | CCTCTGTACAACGCCCAACA | |||
| β-actin-q-F | CATCGGCAATGAGCGGTTCC | 147 | ||
| β-actin-q-R | ACCGTGTTGGCGTAGAGGTC |
| 软件名Software name | 网址 Website | 用途 Purpose |
|---|---|---|
| NCBI | https://www.ncbi.nlm.nih.gov/ | 氨基酸序列 |
| https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi | 保守结构域分析 | |
| DNAMAN9.0软件 | 同源性比对 | |
| MEGA11.0软件 | 构建基因系统进化树 | |
| ExPASy | https://web.expasy.org/protscale/ | 理化性质分析 |
| TMHMM | https://services.healthtech.dtu.dk/service.php?TMHMM-2.0 | 蛋白跨膜结构域预测 |
| SignalP5.0 | https://services.healthtech.dtu.dk/service.php?Signal P-5.0 | 蛋白信号肽预测 |
| NetPhos-3.1 | http://www.cbs.dtu.dk/services/NetPhos/ | 蛋白磷酸化位点分析 |
| NPS@SOPMA | https://npsa-prabi.ibcp.fr/cgi-bin/secpred_sopma.pl | 蛋白质二级结构预测 |
| SWISS-MODEL | https://swissmodel.expasy.org/ | 蛋白质三级结构预测 |
| Meth Primer | http://www.urogene.org/methprimer/ | CpG岛预测 |
| AnimalTFDB4 | bioinfo.life.hust.edu.cn/AnimalTFDB4 | 转录因子结合位点预测 |
| TRANSFAC2.0 | https://genexplain.com/transfac-2-0/ |
Table 2 Software for bioinformatics analysis
| 软件名Software name | 网址 Website | 用途 Purpose |
|---|---|---|
| NCBI | https://www.ncbi.nlm.nih.gov/ | 氨基酸序列 |
| https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi | 保守结构域分析 | |
| DNAMAN9.0软件 | 同源性比对 | |
| MEGA11.0软件 | 构建基因系统进化树 | |
| ExPASy | https://web.expasy.org/protscale/ | 理化性质分析 |
| TMHMM | https://services.healthtech.dtu.dk/service.php?TMHMM-2.0 | 蛋白跨膜结构域预测 |
| SignalP5.0 | https://services.healthtech.dtu.dk/service.php?Signal P-5.0 | 蛋白信号肽预测 |
| NetPhos-3.1 | http://www.cbs.dtu.dk/services/NetPhos/ | 蛋白磷酸化位点分析 |
| NPS@SOPMA | https://npsa-prabi.ibcp.fr/cgi-bin/secpred_sopma.pl | 蛋白质二级结构预测 |
| SWISS-MODEL | https://swissmodel.expasy.org/ | 蛋白质三级结构预测 |
| Meth Primer | http://www.urogene.org/methprimer/ | CpG岛预测 |
| AnimalTFDB4 | bioinfo.life.hust.edu.cn/AnimalTFDB4 | 转录因子结合位点预测 |
| TRANSFAC2.0 | https://genexplain.com/transfac-2-0/ |
| 物种 Species | 登录号 GenBank No. | 氨基酸序列相似性Similarity/% |
|---|---|---|
| 牛 Bos taurus | NP_001039950.2 | |
| 山羊 Capra hircus | XP_005690745.1 | 100 |
| 绵羊 Ovis aries | XP_027831445.1 | 100 |
| 犬 Canis lupus familiaris | XP_038516303.1 | 99.76 |
| 猪 Sus scrofa | XP_020950991.1 | 99.52 |
| 家猫 Felis catus | XP_003989534.1 | 99.03 |
| 人 Homo sapiens | NP_031401.1 | 97.58 |
| 苏门答腊猩猩Pongo abelii | NP_001127597.1 | 97.34 |
| 原鸡 Gallus gallus | NP_001026049.2 | 96.86 |
| 热带爪蟾 Xenopus tropicalis | NP_001231687.1 | 74.10 |
| 小鼠 Mus musculus | NP_001003898.1 | 67.39 |
| 大鼠 Rattus norvegicus | NP_001011979.1 | 66.67 |
Table 3 Amino acid sequence comparison of TARDBP between cattle and different species
| 物种 Species | 登录号 GenBank No. | 氨基酸序列相似性Similarity/% |
|---|---|---|
| 牛 Bos taurus | NP_001039950.2 | |
| 山羊 Capra hircus | XP_005690745.1 | 100 |
| 绵羊 Ovis aries | XP_027831445.1 | 100 |
| 犬 Canis lupus familiaris | XP_038516303.1 | 99.76 |
| 猪 Sus scrofa | XP_020950991.1 | 99.52 |
| 家猫 Felis catus | XP_003989534.1 | 99.03 |
| 人 Homo sapiens | NP_031401.1 | 97.58 |
| 苏门答腊猩猩Pongo abelii | NP_001127597.1 | 97.34 |
| 原鸡 Gallus gallus | NP_001026049.2 | 96.86 |
| 热带爪蟾 Xenopus tropicalis | NP_001231687.1 | 74.10 |
| 小鼠 Mus musculus | NP_001003898.1 | 67.39 |
| 大鼠 Rattus norvegicus | NP_001011979.1 | 66.67 |
| 氨基酸 Amino acid | 数目 Number | 比例Perc-entage/% | 氨基酸 Amino acid | 数目 Number | 比例Perc- entage/% |
|---|---|---|---|---|---|
| Ala(A) | 27 | 6.5% | Leu(L) | 20 | 4.8% |
| Arg(R) | 20 | 4.8% | Lys(K) | 20 | 4.8% |
| Asn(N) | 27 | 6.5% | Met(M) | 17 | 4.1% |
| Asp(D) | 22 | 5.3% | Phe(F) | 22 | 5.3% |
| Cys(C) | 7 | 1.7% | Pro(P) | 17 | 4.1% |
| Gln(Q) | 24 | 5.8% | Ser(S) | 41 | 9.9% |
| Glu(E) | 20 | 4.8% | Thr(T) | 15 | 3.6% |
| Gly(G) | 55 | 13.3% | Trp(W) | 6 | 1.4% |
| His(H) | 5 | 1.2% | Tyr(Y) | 6 | 1.9% |
| Ile(I) | 14 | 3.4% | Val(V) | 8 | 6.5% |
Table 4 Amino acid composition of bovine TARDBP protein
| 氨基酸 Amino acid | 数目 Number | 比例Perc-entage/% | 氨基酸 Amino acid | 数目 Number | 比例Perc- entage/% |
|---|---|---|---|---|---|
| Ala(A) | 27 | 6.5% | Leu(L) | 20 | 4.8% |
| Arg(R) | 20 | 4.8% | Lys(K) | 20 | 4.8% |
| Asn(N) | 27 | 6.5% | Met(M) | 17 | 4.1% |
| Asp(D) | 22 | 5.3% | Phe(F) | 22 | 5.3% |
| Cys(C) | 7 | 1.7% | Pro(P) | 17 | 4.1% |
| Gln(Q) | 24 | 5.8% | Ser(S) | 41 | 9.9% |
| Glu(E) | 20 | 4.8% | Thr(T) | 15 | 3.6% |
| Gly(G) | 55 | 13.3% | Trp(W) | 6 | 1.4% |
| His(H) | 5 | 1.2% | Tyr(Y) | 6 | 1.9% |
| Ile(I) | 14 | 3.4% | Val(V) | 8 | 6.5% |
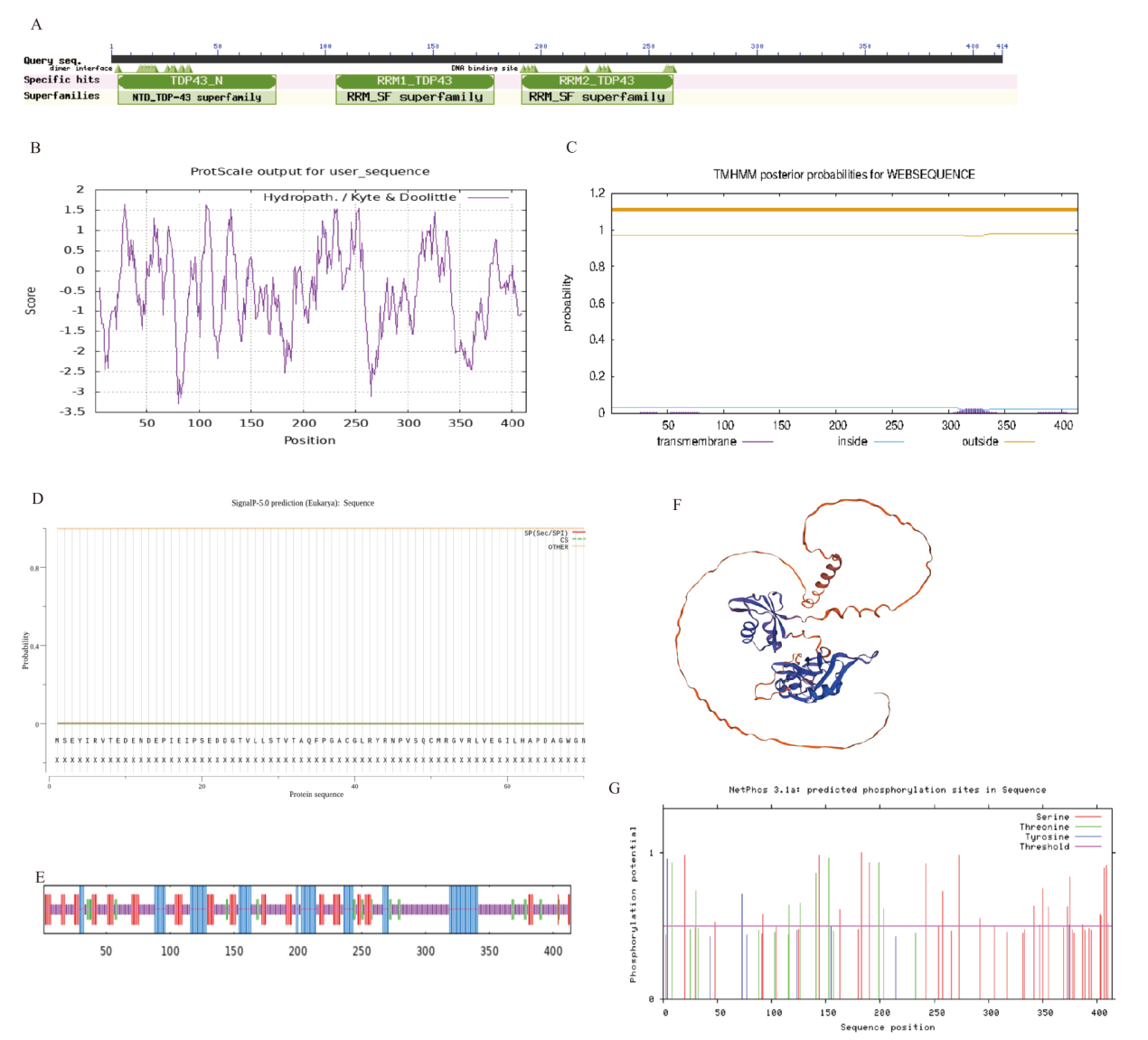
Fig. 3 Analysis of TARDBP protein structure A: Prediction of bovine TARDBP protein conserved domain. B: Prediction of bovine TARDBP protein hydrophobicity. C: Prediction of bovine TARDBP protein transmembrane structure. D: Prediction of bovine TARDBP protein signal peptide. E: Prediction of secondary structure of bovine TARDBP protein. F: Tertiary structure prediction of bovine TARDBP protein. G: Prediction of phosphorylation site of bovine TARDBP protein
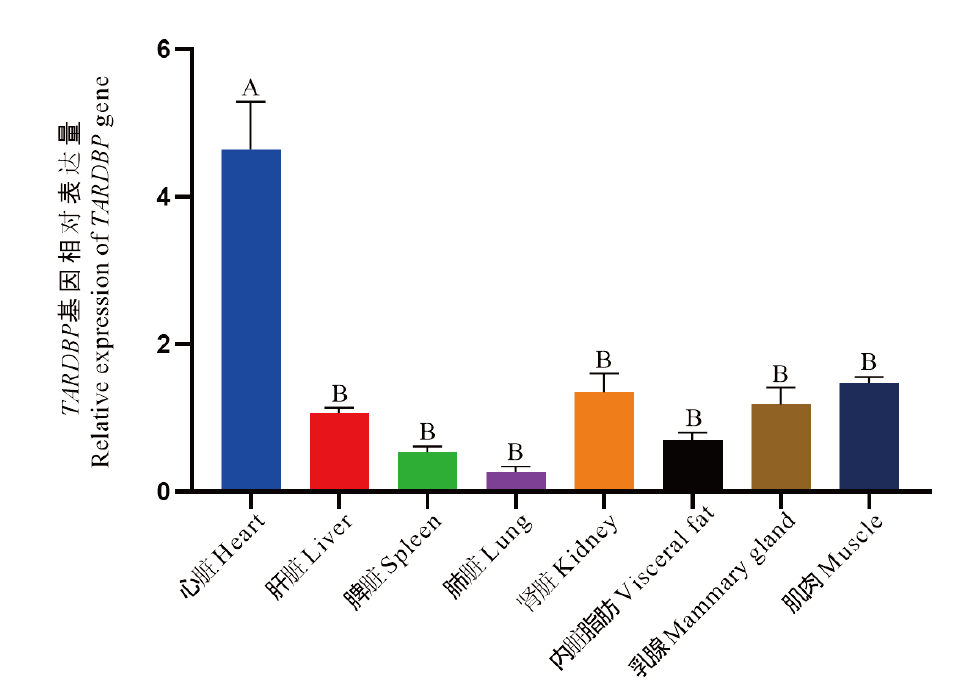
Fig. 4 Analysis of bovine TARDBP gene expression in different tissues The same capital letter indicates that the difference is not significant(P>0.05), different capital letters indicate significant differences(P<0.01)
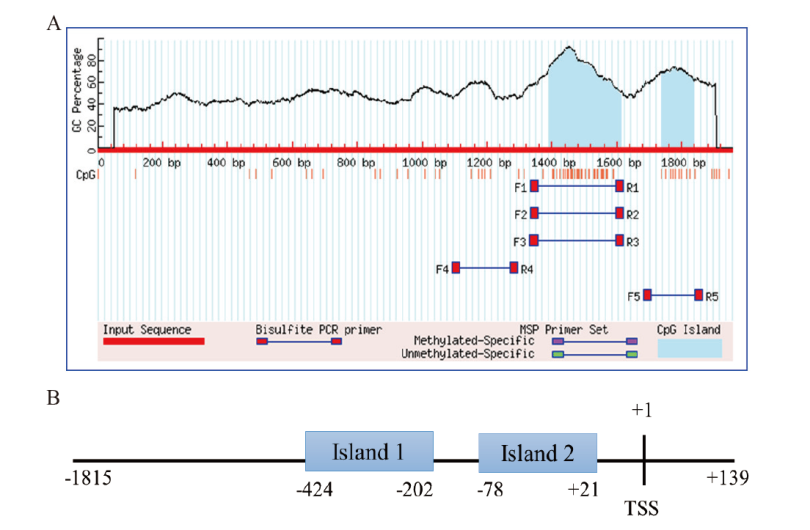
Fig. 8 Prediction results of CpG island in the 5' regulatory region of bovine TARDBP gene A: The predict result by MethPrimer software. B: The specific location of CpG islands

Fig. 9 Prediction of potential transcription factor binding sites of bovine TARDBP gene promoter +1 indicates transcription initiation site; letters in in box indicates transcription factor binding sites. Overlapping regions of transcription factor binding sites are represented by different colored boxes

Fig. 10 Amplification and vector construction of different deletion fragments of bovine TARDBP gene promoter A: Electrophoretic amplification of bovine TARDBP gene promoter fragment by fragment deletion, M: DL5000 DNA marker, 1-6: add the missing fragments of bovine TARDBP gene promoter at double restriction sites. B: Electrophoresis of bovine TARDBP gene promoter double fluorescein reporter recombinant plasmid double enzyme digestion identification, M: DL5000 DNA marker, 1-6: recombinant plasmid
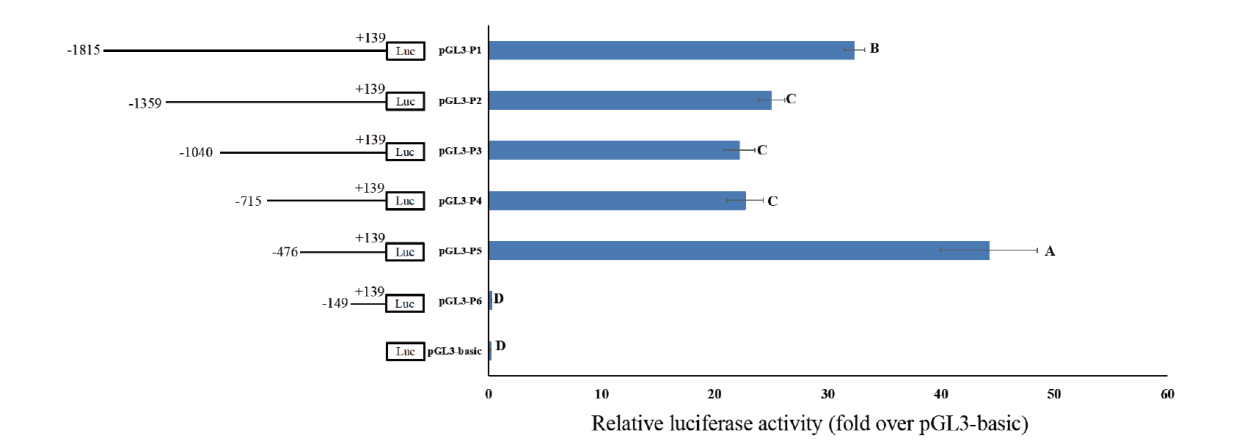
Fig. 11 Activity analysis of bovine TARDBP gene promoter in bovine mammary epithelial cells Different letters indicate significant differences(P<0.05)
| [1] |
Chalupa-Krebzdak S, Long CJ, Bohrer BM. Nutrient density and nutritional value of milk and plant-based milk alternatives[J]. Int Dairy J, 2018, 87: 84-92.
doi: 10.1016/j.idairyj.2018.07.018 URL |
| [2] | Wijesinha-Bettoni R, Burlingame B. Milk and dairy product composition[J]. Milk and dairy products in human nutrition, 2013: 41-102. |
| [3] | Fox PF, Mcsweeney PLH, Paul LH. Dairy chemistry and biochemistry[M]. London: Chapman & Hall,1999. |
| [4] | Samková E, Špička J, Pešek M, et al. Animal factors affecting fatty acid composition of cow milk fat: a review[J]. S Afr N J Anim Sci, 2012, 42(2): 83-100. |
| [5] |
蒋秋斐, 蔡正云, 黄增文, 等. 奶牛乳脂性状候选基因EEF1D突变位点功能分析[J]. 浙江农业学报, 2020, 32(7): 1155-1159.
doi: 10.3969/j.issn.1004-1524.2020.07.03 |
| Jiang QF, Cai ZY, Huang ZW, et al. Functional analysis of EEF1D mutation site in dairy cow milk fat traits candidate gene[J]. Acta Agric Zhejiangensis, 2020, 32(7): 1155-1159. | |
| [6] | Xu B, Gerin I, Miao HZ, et al. Multiple roles for the non-coding RNA SRA in regulation of adipogenesis and insulin sensitivity[J]. PLoS One, 2010, 5(12): e14199. |
| [7] | Chhangani D, Martín-Peña A, Rincon-Limas DE. Molecular, functional, and pathological aspects of TDP-43 fragmentation[J]. iScience, 2021, 24(5): 102459. |
| [8] |
Krecic AM, Swanson MS. hnRNP complexes: composition, structure, and function[J]. Curr Opin Cell Biol, 1999, 11(3): 363-371.
pmid: 10395553 |
| [9] |
Lukavsky PJ, Daujotyte D, Tollervey JR, et al. Molecular basis of UG-rich RNA recognition by the human splicing factor TDP-43[J]. Nat Struct Mol Biol, 2013, 20(12): 1443-1449.
doi: 10.1038/nsmb.2698 pmid: 24240615 |
| [10] |
Weskamp K, Barmada SJ. TDP43 and RNA instability in amyotrophic lateral sclerosis[J]. Brain Res, 2018, 1693(Pt A): 67-74.
doi: S0006-8993(18)30023-4 pmid: 29395044 |
| [11] |
Kuo PH, Chiang CH, Wang YT, et al. The crystal structure of TDP-43 RRM1-DNA complex reveals the specific recognition for UG- and TG-rich nucleic acids[J]. Nucleic Acids Res, 2014, 42(7): 4712-4722.
doi: 10.1093/nar/gkt1407 URL |
| [12] |
Buratti E, Dörk T, Zuccato E, et al. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping[J]. EMBO J, 2001, 20(7): 1774-1784.
doi: 10.1093/emboj/20.7.1774 pmid: 11285240 |
| [13] |
Baralle M, Buratti E, Baralle FE. The role of TDP-43 in the pathogenesis of ALS and FTLD[J]. Biochem Soc Trans, 2013, 41(6): 1536-1540.
doi: 10.1042/BST20130186 URL |
| [14] |
Zhao LM, Li LL, Xu HB, et al. TDP-43 is required for mammary gland repopulation and proliferation of mammary epithelial cells[J]. Stem Cells Dev, 2019, 28(14): 944-953.
doi: 10.1089/scd.2019.0011 pmid: 31062657 |
| [15] | Zhao LM, Ke H, Xu HB, et al. TDP-43 facilitates milk lipid secretion by post-transcriptional regulation of Btn1a1 and Xdh[J]. Nat Commun, 2020, 11(1): 341. |
| [16] | Gowardhan A, Spoon H, Riechers DA, et al. The dual role of starbursts and active galactic nuclei in driving extreme molecular outflows[J]. ApJ, 2018, 859(1): 35. |
| [17] |
Buratti E, Baralle FE. Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease[J]. Front Biosci, 2008, 13: 867-878.
doi: 10.2741/2727 URL |
| [18] |
Warraich ST, Yang S, Nicholson GA, et al. TDP-43: a DNA and RNA binding protein with roles in neurodegenerative diseases[J]. Int J Biochem Cell Biol, 2010, 42(10): 1606-1609.
doi: 10.1016/j.biocel.2010.06.016 pmid: 20601083 |
| [19] |
Stover CM, Lynch NJ, Hanson SJ, et al. Organization of the MASP2 locus and its expression profile in mouse and rat[J]. Mamm Genome, 2004, 15(11): 887-900.
pmid: 15672593 |
| [20] |
Ou SH, Wu F, Harrich D, et al. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs[J]. J Virol, 1995, 69(6): 3584-3596.
doi: 10.1128/JVI.69.6.3584-3596.1995 pmid: 7745706 |
| [21] | 郑敏. 家猪全基因组甲基化时空变化以及重复元件PRE1甲基化功能研究[D]. 南昌: 江西农业大学, 2022. |
| Zheng M. Study on the spatiotemporal variations of pig methylomes and the function of methylated PRE1[D]. Nanchang: Jiangxi Agricultural University, 2022. | |
| [22] | Oh YM, Mahar M, Ewan EE, et al. Epigenetic regulator UHRF1 inactivates REST and growth suppressor gene expression via DNA methylation to promote axon regeneration[J]. Proc Natl Acad Sci USA, 2018, 115(52): E12417-E12426. |
| [23] | Xiang Y, Chen QX, Li QB, et al. The expression level of chicken telomerase reverse transcriptase in tumors induced by ALV-J is positively correlated with methylation and mutation of its promoter region[J]. Vet Res, 2022, 53(1): 49. |
| [24] |
Lenhard B, Sandelin A, Carninci P. Metazoan promoters: emerging characteristics and insights into transcriptional regulation[J]. Nat Rev Genet, 2012, 13(4): 233-245.
doi: 10.1038/nrg3163 pmid: 22392219 |
| [25] |
Kadonaga JT, Carner KR, Masiarz FR, et al. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain[J]. Cell, 1987, 51(6): 1079-1090.
doi: 10.1016/0092-8674(87)90594-0 pmid: 3319186 |
| [26] |
Song J, Ugai H, Nakata-Tsutsui H, et al. Transcriptional regulation by zinc-finger proteins Sp1 and MAZ involves interactions with the same cis-elements[J]. Int J Mol Med, 2003, 11(5): 547-553.
pmid: 12684688 |
| [27] | 杨洋, 周子薇, 张京一, 等. 奶牛SP1基因结构及对乳脂合成功能的初步分析[J]. 畜牧兽医学报, 2022, 53(9): 2970-2981. |
| Yang Y, Zhou ZW, Zhang JY, et al. Analysison SP1 gene structure and Its functionon milk fat synthesisin holstein dairy cows[J]. Acta Veterinaria Et Zootechnica Sinica, 2022, 53(9):2970-2981. | |
| [28] |
Zhu JJ, Luo J, Xu HF, et al. Short communication: altered expression of specificity protein 1 impairs milk fat synthesis in goat mammary epithelial cells[J]. J Dairy Sci, 2016, 99(6): 4893-4898.
doi: S0022-0302(16)30086-8 pmid: 26995134 |
| [29] |
Bionaz M, Loor JJ. Gene networks driving bovine milk fat synthesis during the lactation cycle[J]. BMC Genomics, 2008, 9: 366.
doi: 10.1186/1471-2164-9-366 pmid: 18671863 |
| [30] |
Yu K, Shi HB, Luo J, et al. PPARG modulated lipid accumulation in dairy GMEC via regulation of ADRP gene[J]. J Cell Biochem, 2015, 116(1): 192-201.
doi: 10.1002/jcb.v116.1 URL |
| [31] |
Kadegowda AKG, Bionaz M, Piperova LS, et al. Peroxisome proliferator-activated receptor-γ activation and long-chain fatty acids alter lipogenic gene networks in bovine mammary epithelial cells to various extents[J]. J Dairy Sci, 2009, 92(9): 4276-4289.
doi: 10.3168/jds.2008-1932 pmid: 19700688 |
| [32] |
Shi HB, Luo J, Yao DW, et al. Peroxisome proliferator-activated receptor-γ stimulates the synthesis of monounsaturated fatty acids in dairy goat mammary epithelial cells via the control of stearoyl-coenzyme A desaturase[J]. J Dairy Sci, 2013, 96(12): 7844-7853.
doi: 10.3168/jds.2013-7105 URL pmid: 24119817 |
| [33] |
Shi HB, Zhao WS, Luo J, et al. Peroxisome proliferator-activated receptor γ1 and γ2 isoforms alter lipogenic gene networks in goat mammary epithelial cells to different extents[J]. J Dairy Sci, 2014, 97(9): 5437-5447.
doi: 10.3168/jds.2013-7863 pmid: 25022676 |
| [34] |
Shi HB, Zhang CH, Xu ZA, et al. Peroxisome proliferator-activated receptor delta regulates lipid droplet formation and transport in goat mammary epithelial cells[J]. J Dairy Sci, 2018, 101(3): 2641-2649.
doi: S0022-0302(18)30014-6 pmid: 29331469 |
| [35] |
Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor[J]. Cell, 1997, 89(3): 331-340.
doi: 10.1016/s0092-8674(00)80213-5 pmid: 9150132 |
| [36] | Zhao X, Li J, Zhao SY, et al. Regulation of bta-miRNA29d-3p on lipid accumulation via GPAM in bovine mammary epithelial cells[J]. Agriculture, 2023, 13(2): 501. |
| [37] |
Jeon TI, Osborne TF. SREBPs: metabolic integrators in physiology and metabolism[J]. Trends Endocrinol Metab, 2012, 23(2): 65-72.
doi: 10.1016/j.tem.2011.10.004 URL |
| [38] |
Cheng X, Li JY, Guo DL. SCAP/SREBPs are central players in lipid metabolism and novel metabolic targets in cancer therapy[J]. Curr Top Med Chem, 2018, 18(6): 484-493.
doi: 10.2174/1568026618666180523104541 pmid: 29788888 |
| [39] |
Xue LY, Qi HY, Zhang H, et al. Targeting SREBP-2-regulated mevalonate metabolism for cancer therapy[J]. Front Oncol, 2020, 10: 1510.
doi: 10.3389/fonc.2020.01510 pmid: 32974183 |
| [40] |
Xu HF, Luo J, Zhao WS, et al. Overexpression of SREBP1(sterol regulatory element binding protein 1)promotes de novo fatty acid synthesis and triacylglycerol accumulation in goat mammary epithelial cells[J]. J Dairy Sci, 2016, 99(1): 783-795.
doi: 10.3168/jds.2015-9736 pmid: 26601584 |
| [1] | XIAO Ya-ru, JIA Ting-ting, LUO Dan, WU Zhe, LI Li-xia. Cloning and Expression Analysis of CsERF025L Transcription Factor in Cucumber [J]. Biotechnology Bulletin, 2024, 40(4): 159-166. |
| [2] | ZHONG Yun, LIN Chun, LIU Zheng-jie, DONG Chen-wen-hua, MAO Zi-chao, LI Xing-yu. Cloning and Prokaryotic Expression Analysis of Asparagus Saponin Synthesis Related Glycosyltransferase Genes [J]. Biotechnology Bulletin, 2024, 40(4): 255-263. |
| [3] | CHEN Xiao-song, LIU Chao-jie, ZHENG Jia, QIAO Zong-wei, LUO Hui-bo, ZOU Wei. Analyzing the Growth and Caproic Acid Metabolism Mechanism of Rummeliibacillus suwonensis 3B-1 by Tandem Mass Tag-based Quantitative Proteomics [J]. Biotechnology Bulletin, 2024, 40(3): 135-145. |
| [4] | YANG Wei-jie, YANG Zhou-lin, ZHU Hao-dong, WEI Yu, LIU Jun, LIU Xun. Study on the Properties and Functions of LchAD Protein, a Key Module of Lichenysin Synthase [J]. Biotechnology Bulletin, 2024, 40(3): 322-332. |
| [5] | GONG Li-li, YU Hua, YANG Jie, CHEN Tian-chi, ZHAO Shuang-ying, WU Yue-yan. Identification and Analysis of Grape(Vitis vinifera L.)CYP707A Gene Family and Functional Verification to Fruit Ripening [J]. Biotechnology Bulletin, 2024, 40(2): 160-171. |
| [6] | ZHU Yi, LIU Tang-jing, GONG Guo-yi, ZHANG Jie, WANG Jin-fang, ZHANG Hai-ying. Cloning and Expression Analysis of ClPP2C3 in Citrullus lanatus [J]. Biotechnology Bulletin, 2024, 40(1): 243-249. |
| [7] | LIU Yu-ling, WANG Meng-yao, SUN Qi, MA Li-hua, ZHU Xin-xia. Effect of RD29A Promoter on the Stress Resistance of Transgenic Tobacco with SikCDPK1 Gene from Saussurea involucrata [J]. Biotechnology Bulletin, 2023, 39(9): 168-175. |
| [8] | ZHANG Lu-yang, HAN Wen-long, XU Xiao-wen, YAO Jian, LI Fang-fang, TIAN Xiao-yuan, ZHANG Zhi-qiang. Identification and Expression Analysis of the Tobacco TCP Gene Family [J]. Biotechnology Bulletin, 2023, 39(6): 248-258. |
| [9] | LI Jing-rui, WANG Yu-bo, XIE Zi-wei, LI Chang, WU Xiao-lei, GONG Bin-bin, GAO Hong-bo. Identification and Expression Analysis of PIN Gene Family in Melon Under High Temperature Stress [J]. Biotechnology Bulletin, 2023, 39(5): 192-204. |
| [10] | GUO San-bao, SONG Mei-ling, LI Ling-xin, YAO Zi-zhao, GUI Ming-ming, HUANG Sheng-he. Cloning and Analysis of Chalcone Synthase Gene and Its Promoter from Euphorbia maculata [J]. Biotechnology Bulletin, 2023, 39(4): 148-156. |
| [11] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [12] | YANG Lan, ZHANG Chen-xi, FAN Xue-wei, WANG Yang-guang, WANG Chun-xiu, LI Wen-ting. Gene Cloning, Expression Pattern, and Promoter Activity Analysis of Chicken BMP15 [J]. Biotechnology Bulletin, 2023, 39(4): 304-312. |
| [13] | CHEN Qiang, ZHOU Ming-kang, SONG Jia-min, ZHANG Chong, WU Long-kun. Identification and Analysis of LBD Gene Family and Expression Analysis of Fruit Development in Cucumis melo [J]. Biotechnology Bulletin, 2023, 39(3): 176-183. |
| [14] | PING Huai-lei, GUO Xue, YU Xiao, SONG Jing, DU Chun, WANG Juan, ZHANG Huai-bi. Cloning and Expression of PdANS in Paeonia delavayi and Correlation with Anthocyanin Content [J]. Biotechnology Bulletin, 2023, 39(3): 206-217. |
| [15] | XING Yuan, SONG Jian, LI Jun-yi, ZHENG Ting-ting, LIU Si-chen, QIAO Zhi-jun. Identification of AP Gene Family and Its Response Analysis to Abiotic Stress in Setaria italica [J]. Biotechnology Bulletin, 2023, 39(11): 238-251. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||