








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (9): 92-103.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0528
Previous Articles Next Articles
QIAO Yan1,3( ), YANG Fang1,2, REN Pan-rong1,2, QI Wei-liang1,2, AN Pei-pei1,2, LI Qian1,2, LI Dan1,2, XIAO Jun-fei4
), YANG Fang1,2, REN Pan-rong1,2, QI Wei-liang1,2, AN Pei-pei1,2, LI Qian1,2, LI Dan1,2, XIAO Jun-fei4
Received:2024-05-28
Online:2024-09-26
Published:2024-10-12
Contact:
QIAO Yan
E-mail:yanqiao@ldxy.edu.cn
QIAO Yan, YANG Fang, REN Pan-rong, QI Wei-liang, AN Pei-pei, LI Qian, LI Dan, XIAO Jun-fei. Cloning and Function Analysis of the ScDHNS Gene of Crotonase/Enoyl-CoA Superfamily from a Wild Potato Species[J]. Biotechnology Bulletin, 2024, 40(9): 92-103.
| 引 物Primer | 序列Sequence(5'-3') |
|---|---|
| P1(g21236-5GSP1) | ACATTAAGGCGACCAAAACTTTCA |
| P2(g21236-5GSP2) | AACTTTCAAAATCAGCATAACCATCC |
| P3(outer primer) | GCTGTCAACGATACGCTACGTAAC |
| P4(inner primer) | GCTACGTAACGGCATGACAGTG |
| P5(g21236-3GSP1) | CCATTAGATAAGTTGGAGGCAG |
| P6(g21236-3GSP2) | GTCCTACAGCGATACGAGTGC |
| P7(outer primer) | TACCGTCGTTCCACTAGTGATTT |
| P8(inner primer) | CGCGGATCCTCCACTAGTGATTTCACTATAGG |
Table 1 Primers for the 5'- and 3'- RACE analysis
| 引 物Primer | 序列Sequence(5'-3') |
|---|---|
| P1(g21236-5GSP1) | ACATTAAGGCGACCAAAACTTTCA |
| P2(g21236-5GSP2) | AACTTTCAAAATCAGCATAACCATCC |
| P3(outer primer) | GCTGTCAACGATACGCTACGTAAC |
| P4(inner primer) | GCTACGTAACGGCATGACAGTG |
| P5(g21236-3GSP1) | CCATTAGATAAGTTGGAGGCAG |
| P6(g21236-3GSP2) | GTCCTACAGCGATACGAGTGC |
| P7(outer primer) | TACCGTCGTTCCACTAGTGATTT |
| P8(inner primer) | CGCGGATCCTCCACTAGTGATTTCACTATAGG |
| 引物名称Primer name | 引物序列 Primer sequence(5'-3') | 退火温度 Annealing temperature/℃ | 长度 Length/bp |
|---|---|---|---|
| SGT1-F | AAGCCACAATCCTCACTACCC | 57.7 | 132 |
| SGT1-R | AGGCAACCCAACTTCAGCAG | 59.2 | |
| ScDHNS-F | TTGATGATGGACATGCTGGACTTC | 58.0 | 84 |
| ScDHNS-R | TGCCTTCATTGCCTTCTTCAGTTC | 59.0 | |
| ef1-α-F | ATTCAAGTATGCCTGGGTGCT | 58.9 | 144 |
| ef1-α-R | TTCTTGATAAAGTCTCTGTGTCCG | 58.4 |
Table 2 Primers’ sequences for RT-qPCR
| 引物名称Primer name | 引物序列 Primer sequence(5'-3') | 退火温度 Annealing temperature/℃ | 长度 Length/bp |
|---|---|---|---|
| SGT1-F | AAGCCACAATCCTCACTACCC | 57.7 | 132 |
| SGT1-R | AGGCAACCCAACTTCAGCAG | 59.2 | |
| ScDHNS-F | TTGATGATGGACATGCTGGACTTC | 58.0 | 84 |
| ScDHNS-R | TGCCTTCATTGCCTTCTTCAGTTC | 59.0 | |
| ef1-α-F | ATTCAAGTATGCCTGGGTGCT | 58.9 | 144 |
| ef1-α-R | TTCTTGATAAAGTCTCTGTGTCCG | 58.4 |
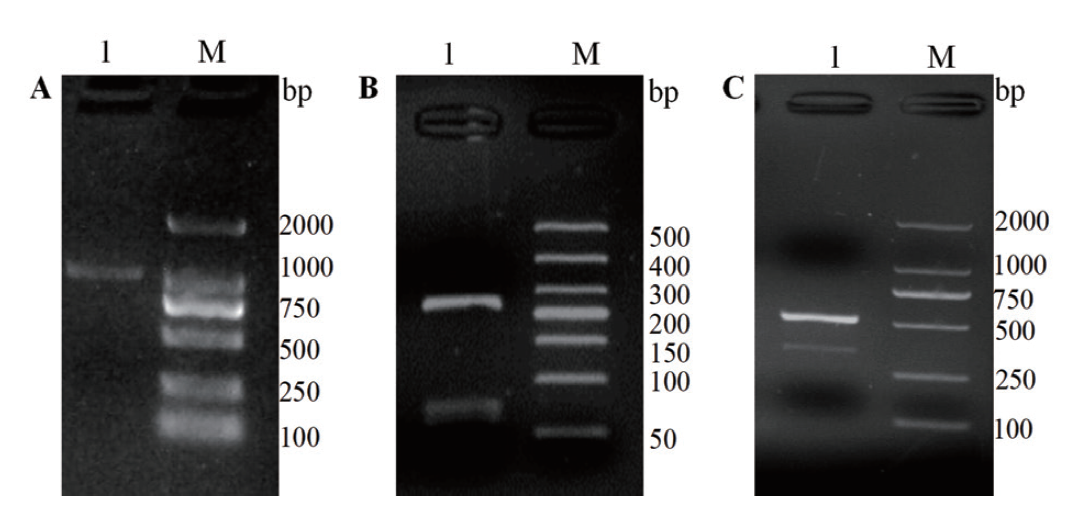
Fig. 1 The core fragment, 5'- and 3'- end fragments of gene ScDHNS A: The core fragment of ScDHNS gene(Lane M: 2000 DNA marker; lane 1: CDS cloning of ScDHNS gene). B: 5'-RACE end fragments of ScDHNS gene(Lane M: 500 DNA marker. Lane 1: 5'- RACE fragments of ScDHNS gene). C: 3'-RACE fragments of ScDHNS gene(Lane M: 2000 DNA marker. Lane 1: 5'- RACE fragments of ScDHNS gene)

Fig. 2 Nucleotide sequences and deduced amino acid sequences of gene ScDHNS The black boxes indicate the initiation and termination codons, respectively. Asterisk indicates the stop codon

Fig. 3 Multiple comparisons of amino acid sequences between ScDHNS and other plants The ECH domain is displayed as line. β-strands are rendered as arrows, strict β-turns as TT letters. The helix line indicates the α helix or 310- helix(η)area. DHNS gene sources: ScDHNS(Solanum chacoense), SlDHNS(Solanum lycopersicum), StDHNS(Solanum tuberosum), NaDHNS(Nicotiana attenuata), BdDHNS(Brachypodium distachyon), AtDHNS(Arabidopsis thaliana)and MtDHNS(Medicago truncatula)
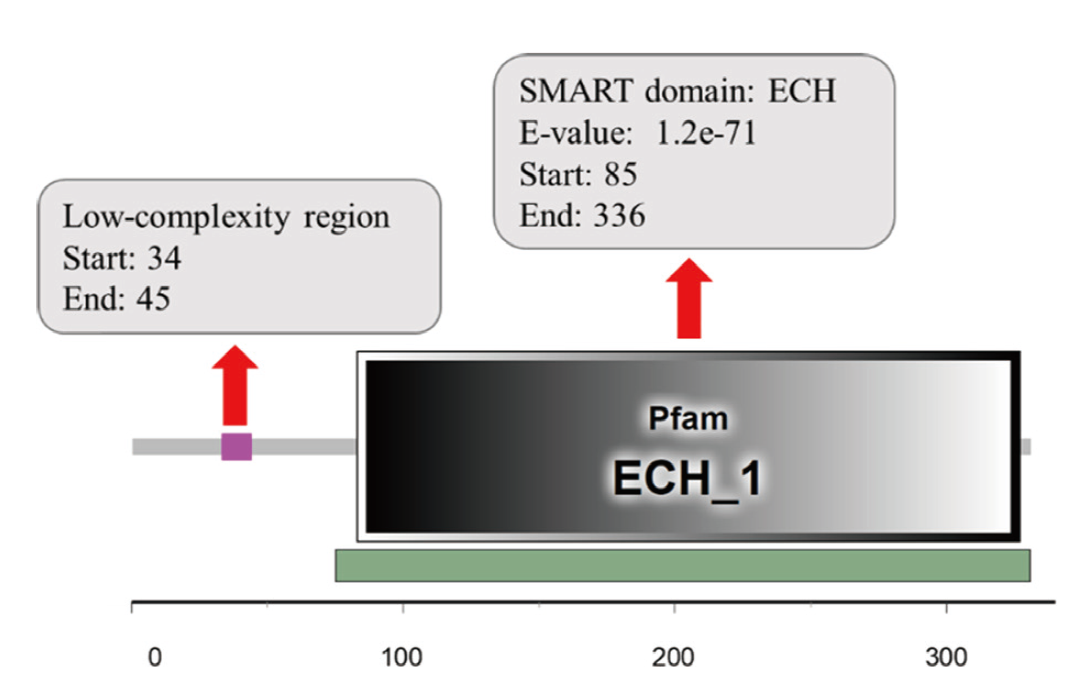
Fig. 4 Functional domain analysis of ScDHNS protein SMART domain indicates the predicted structural domain of ScDHNS. The scale indicates the order of the codon starting from the initial codon
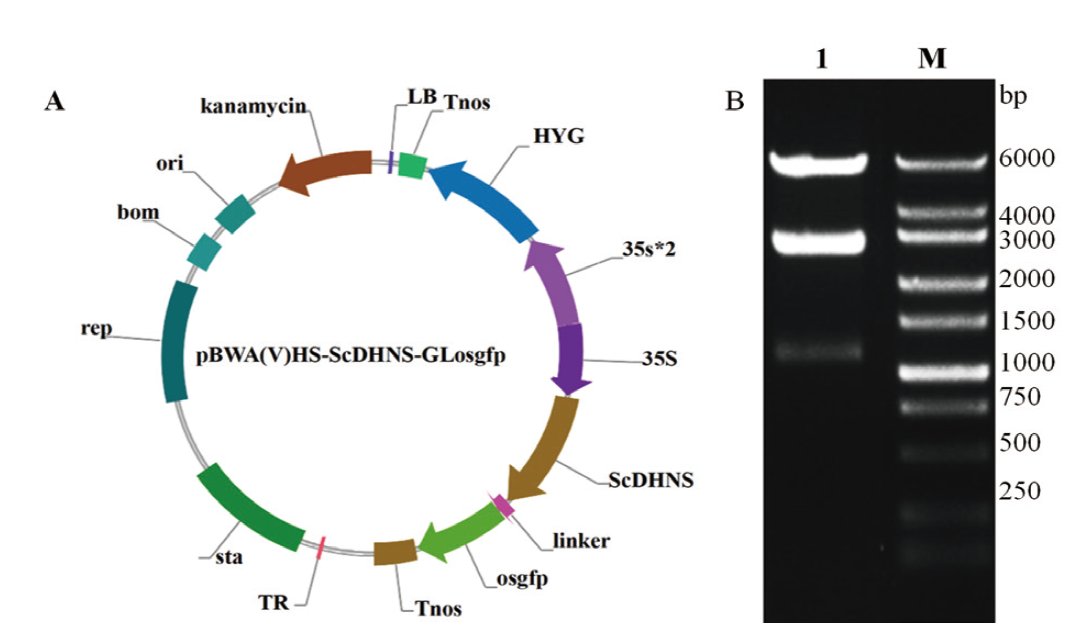
Fig. 6 Profile of pBWA-ScDHNS-Glosgfp subcellular localization vector and its digestion verification A: Profile of pBWA-ScDHNS-Glosgfp subcellular localization vector. B: Digestion result of pBWA-ScDHNS-Glosgfp vector(Lane M: DNA marker. Lane 1: Digestion result of vector)

Fig. 7 Subcellular localization of ScDHNS in Arabidopsis protoplasts A: Empty vector pBWA-Glosgfp was transformed into Arabidopsis protoplasts. B: Recombinant vector pBWA(V)HS-ScDHNS-Glosgfp was transformed into Arabidopsis protoplasts
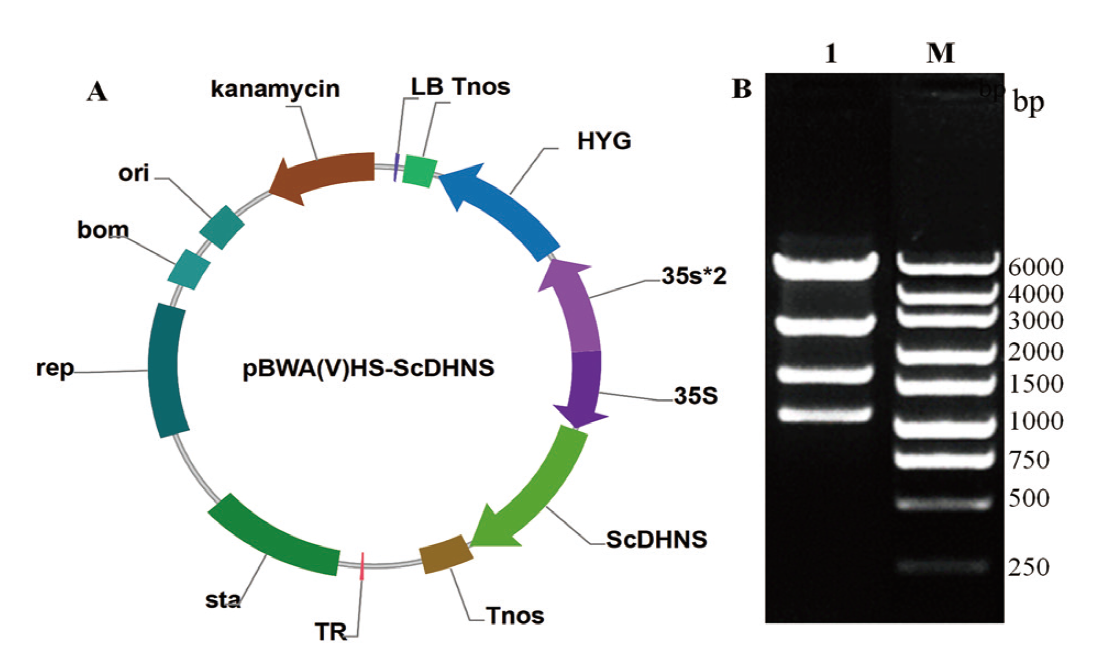
Fig. 8 Profile of pBWA(V)HS-ScDHNS expressing vector and its enzymatic digestion verification A: pBWA(V)HS-ScDHNS express vector. B: Digestion result of pBWA(V)HS-ScDHNS vector(Lane M: DNA marker. Lane 1: Digestion result of vector)
| [1] | 赵锋, 胡开明, 王晓斌, 等. 饲用马铃薯潜在产量的分析方法[J]. 草业科学, 2016, 33(11): 2326-2336. |
| Zhao F, Hu KM, Wang XB, et al. Analysis method of forage-use potato potential yields[J]. Pratacultural Sci, 2016, 33(11): 2326-2336. | |
| [2] | 夏善勇. 马铃薯渣与茎叶饲料化利用及研究进展[J]. 中国饲料, 2023(17): 137-143. |
| Xia SY. Research progress on forage utilization of potato residue and vine[J]. China Feed, 2023(17): 137-143. | |
| [3] | Cao QH, Dai WF, Li BC, et al. Sesquiterpenoids from the stems and leaves of Gochnatia decora[J]. Phytochem Lett, 2019, 30: 6-9. |
| [4] | 雒瑞瑞. 马铃薯茎叶和玉米秸/甜高粱混合青贮料的制作及其对瘤胃发酵特性的影响[D]. 兰州: 甘肃农业大学, 2018. |
| Luo RR. Preparation of mixed silage of potato stem and leaf and corn stalk/sweet sorghum and its influence on rumen fermentation characteristics[D]. Lanzhou: Gansu Agricultural University, 2018. | |
| [5] | Zhang SY, Deng JJ, Cui YF, et al. Effect of potato vine and leaf mixed silage to whole corn crops on rumen fermentation and the microbe of fatten Angus bulls[J]. Fermentation, 2023, 9(8): 704. |
| [6] | Qiao Y, Yang F, Li Q, et al. Combined small RNA and degradome sequencing reveals important roles of light-responsive microRNAs in wild potato(Solanum chacoense)[J]. Agronomy, 2023, 13(7): 1763. |
| [7] | Sakhare AV, Bess S, Mill LL. Isolation of solanine from potato leaves and evaluation of its antimicrobial activity[J]. Int J Sci Res, 2014, 3(11): 2052-2056. |
| [8] | Nützmann HW, Huang AC, Osbourn A. Plant metabolic clusters - from genetics to genomics[J]. New Phytol, 2016, 211(3): 771-789. |
| [9] | Zhao DK, Zhao Y, Chen SY, et al. Solanum steroidal glycoalkaloids: structural diversity, biological activities, and biosynthesis[J]. Nat Prod Rep, 2021, 38(8): 1423-1444. |
| [10] | Sonawane PD, Jozwiak A, Panda S, et al. ‘Hijacking’ core metabolism: a new panache for the evolution of steroidal glycoalkaloids structural diversity[J]. Curr Opin Plant Biol, 2020, 55: 118-128. |
| [11] |
Itkin M, Heinig U, Tzfadia O, et al. Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes[J]. Science, 2013, 341(6142): 175-179.
doi: 10.1126/science.1240230 pmid: 23788733 |
| [12] |
Cárdenas PD, Sonawane PD, Pollier J, et al. GAME9 regulates the biosynthesis of steroidal alkaloids and upstream isoprenoids in the plant mevalonate pathway[J]. Nat Commun, 2016, 7: 10654.
doi: 10.1038/ncomms10654 pmid: 26876023 |
| [13] | McCue KF, Shepherd LVT, Allen PV, et al. Metabolic compensation of steroidal glycoalkaloid biosynthesis in transgenic potato tubers: using reverse genetics to confirm the in vivo enzyme function of a steroidal alkaloid galactosyltransferase[J]. Plant Sci, 2005, 168(1): 267-273. |
| [14] | Widhalm JR, Ducluzeau AL, Buller NE, et al. Phylloquinone(vitamin K(1))biosynthesis in plants: two peroxisomal thioesterases of Lactobacillales origin hydrolyze 1, 4-dihydroxy-2-naphthoyl-CoA[J]. Plant J, 2012, 71(2): 205-215. |
| [15] | Kumar S, Tripathi J, Srivastava AK, et al. Molecular mechanism of antimutagenicity by an ethoxy-substituted phylloquinone(vitamin K1 derivative)from spinach(Spinacea oleracea L.)[J]. Chem Biol Interact, 2020, 330: 109216. |
| [16] |
Mweetwa AM, Hunter D, Poe R, et al. Steroidal glycoalkaloids in Solanum chacoense[J]. Phytochemistry, 2012, 75: 32-40.
doi: 10.1016/j.phytochem.2011.12.003 pmid: 22217745 |
| [17] |
Schaefer BC. Revolutions in rapid amplification of cDNA ends: new strategies for polymerase chain reaction cloning of full-length cDNA ends[J]. Anal Biochem, 1995, 227(2): 255-273.
pmid: 7573945 |
| [18] | Beaujean A, Sangwan RS, Lecardonnel A, et al. Agrobacterium -mediated transformation of three economically important potato cultivars using sliced internodal explants: an efficient protocol of transformation[J]. J Exp Bot, 1998, 49(326): 1589-1595. |
| [19] | Grunenfelder LA, Knowles LO, Hiller LK, et al. Glycoalkaloid development during greening of fresh market potatoes(Solanum tuberosum L.)[J]. J Agric Food Chem, 2006, 54(16): 5847-5854. |
| [20] | Bolser DM, Kerhornou A, Walts B, et al. Triticeae resources in Ensembl Plants[J]. Plant Cell Physiol, 2015, 56(1): e3. |
| [21] |
De Bie T, Cristianini N, Demuth JP, et al. CAFE: a computational tool for the study of gene family evolution[J]. Bioinformatics, 2006, 22(10): 1269-1271.
doi: 10.1093/bioinformatics/btl097 pmid: 16543274 |
| [22] | Chou KC, Shen HB. Cell-PLoc: a package of Web servers for predicting subcellular localization of proteins in various organisms[J]. Nat Protoc, 2008, 3(2): 153-162. |
| [23] | 乔岩. 马铃薯光诱导糖苷生物碱代谢相关miRNAs的鉴定与功能分析[D]. 兰州: 甘肃农业大学, 2017. |
| Qiao Y. Identification and functional analysis of miRNAs related to photoinduced glycoside alkaloid metabolism in potato[D]. Lanzhou: Gansu Agricultural University, 2017. | |
| [24] | Huang J, Xu WB, Zhai JW, et al. Nuclear phylogeny and insights into whole-genome duplications and reproductive development of Solanaceae plants[J]. Plant Commun, 2023, 4(4): 100595. |
| [25] | Dalwani S, Wierenga RK. Enzymes of the crotonase superfamily: diverse assembly and diverse function[J]. Curr Opin Struct Biol, 2023, 82: 102671. |
| [26] | Greenhagen BT, Schoenbeck MA, Yeo YS, et al. Chapter ten The chemical wizardry of isoprenoid metabolism in plants[M]// Recent Advances in Phytochemistry. Amsterdam: Elsevier, 2003: 231-251. |
| [27] | del Río LA, Schrader M. Proteomics of peroxisomes: identifying novel functions and regulatory networks[M]. Singapore: Springer Singapore, 2018:3-45. |
| [28] |
Pan RH, Liu J, Wang SS, et al. Peroxisomes: versatile organelles with diverse roles in plants[J]. New Phytol, 2020, 225(4): 1410-1427.
doi: 10.1111/nph.16134 pmid: 31442305 |
| [29] |
Li HB, Brouwer M, Pup ED, et al. Allelic variation in the autotetraploid potato: genes involved in starch and steroidal glycoalkaloid metabolism as a case study[J]. BMC Genomics, 2024, 25(1): 274.
doi: 10.1186/s12864-024-10186-5 pmid: 38475714 |
| [1] | XING Li-nan, ZHANG Yan-fang, GE Ming-ran, ZHAO Ling-min, CHEN Yan, HUO Xiu-wen. Analysis of DoWRKY40 Gene Expression Characteristics and Screening of Interacting Proteins in Yam [J]. Biotechnology Bulletin, 2024, 40(8): 118-128. |
| [2] | LIN Tong, YUAN Cheng, DONG Chen-wen-hua, ZENG Meng-qiong, YANG Yan, MAO Zi-chao, LIN Chun. Screening and Functional Analysis of Gene CqSTK Associated with Gametophyte Development of Quinoa [J]. Biotechnology Bulletin, 2024, 40(8): 83-94. |
| [3] | SHEN Zhen-hui, CAO Yao, YANG Lin-lei, LUO Xiang-ying, ZI Ling-shan, LU Qing-qing, LI Rong-chun. Cloning and Bioinformatics Analysis of the Ergothioneine Biosynthesis Genes in Naematelia aurantialba and Stereum hirsutum [J]. Biotechnology Bulletin, 2024, 40(7): 259-272. |
| [4] | HUANG Dan, JIANG Shan, PENG Tao. Cloning of FfCYP98 Gene and Its Functional Analysis in Folioceros fuciformis [J]. Biotechnology Bulletin, 2024, 40(7): 273-284. |
| [5] | PANG Meng-zhen, XU Han-qin, LIU Hai-yan, SONG Juan, WANG Jia-han, SUN Li-na, JI Pei-mei, YIN Ze-zhi, HU You-chuan, ZHAO Xiao-meng, LIANG Shan-shan, ZHANG Si-ju, LUAN Wei-jiang. Gene Identification and Functional Analysis of Yellowish and Early Heading Mutant hz1 in Rice [J]. Biotechnology Bulletin, 2024, 40(7): 125-136. |
| [6] | LI Bo-jing, ZHENG La-mei, WU Wu-yun, GAO Fei, ZHOU Yi-jun. Evolution, Expression, and Functional Analysis of the HSP20 Gene Family from Simmondisa chinensis [J]. Biotechnology Bulletin, 2024, 40(6): 190-202. |
| [7] | WU Ze-hang, YANG Zhong-yi, YAN Yi-cheng, JIA Yong-hong, WU Yue-yan, XIE Xiao-hong. Cloning and Functional Analysis of Flavonoid 3'-hydroxylase(F3'H)Gene in Rhododendron hybridum Hort [J]. Biotechnology Bulletin, 2024, 40(6): 251-259. |
| [8] | YAN Huan-huan, SHANG Yi-tong, WANG Li-hong, TIAN Xue-qin, LIAO Hai-yan, ZENG Bin, HU Zhi-hong. Heterologous Biosynthesis of Cordycepin in Aspergillus oryzae [J]. Biotechnology Bulletin, 2024, 40(6): 290-298. |
| [9] | WANG Yu-shu, ZHAO Lin-lin, ZHAO Shuang, HU Qi, BAI Hui-xia, WANG Huan, CAO Ye-ping, FAN Zhen-yu. Cloning and Expression Analysis of BrCYP83B1 Gene in Chinese Cabbage [J]. Biotechnology Bulletin, 2024, 40(6): 152-160. |
| [10] | HAO Si-yi, ZHANG Jun-ke, WANG Bin, QU Peng-yan, LI Rui-de, CHENG Chun-zhen. Cloning and Expression Analysis of Banana EARLY FLOWERING 3(ELF3)Genes [J]. Biotechnology Bulletin, 2024, 40(5): 131-140. |
| [11] | PAN Ping-ping, XU Zhi-hao, ZHANG Yi-wen, LI Qing, WANG Zhong-hua. Prokaryotic Expression, Subcellular Localization and Expression Analysis of PcCHS Gene from Polygonatum cyrtonema Hua [J]. Biotechnology Bulletin, 2024, 40(5): 280-289. |
| [12] | ZHANG Zhen, LI Qing, XU Jing, CHEN Kai-yuan, ZHANG Chun-zhi, ZHU Guang-tao. Construction and Application of Potato Mitochondrial Targeted Expression Vector [J]. Biotechnology Bulletin, 2024, 40(5): 66-73. |
| [13] | DU Ze-guang, REN Shao-wen, ZHANG Feng-qin, LI Mei-lan, LI Gai-zhen, QI Xian-hui. Cloning,Expression and Functional Identification of BrMLP328 Gene in Brassica rapa subsp. pekinensis [J]. Biotechnology Bulletin, 2024, 40(4): 122-129. |
| [14] | LIU Huan-huan, YANG Li-chun, LI Huo-gen. Cloning and Functional Analysis of LtMYB305 in Liriodendron tulipifera [J]. Biotechnology Bulletin, 2024, 40(4): 179-188. |
| [15] | ZHONG Yun, LIN Chun, LIU Zheng-jie, DONG Chen-wen-hua, MAO Zi-chao, LI Xing-yu. Cloning and Prokaryotic Expression Analysis of Asparagus Saponin Synthesis Related Glycosyltransferase Genes [J]. Biotechnology Bulletin, 2024, 40(4): 255-263. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||