








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (3): 104-111.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0839
MA Li-hua( ), HOU Meng-juan, ZHU Xin-xia(
), HOU Meng-juan, ZHU Xin-xia( )
)
Received:2024-08-29
Online:2025-03-26
Published:2025-03-20
Contact:
ZHU Xin-xia
E-mail:2271579422@qq.com;302641316@qq.com
MA Li-hua, HOU Meng-juan, ZHU Xin-xia. Functional Studies of the Gossypium hirsutumGhNFD4 Gene in Response to Drought in Cotton[J]. Biotechnology Bulletin, 2025, 41(3): 104-111.
引物名称 Primer name | 引物序列 Primer sequence (5′-3′) | 用途Purpose |
|---|---|---|
| GhUBQ7-F | AGAGGTCGAGTCTTGGGACA | 内参基因Reference gene |
| GhUBQ7-R | GCTTGATCTTCTTGGGCTTG | |
| GhNFD4-F | AAACCTTGTCACTTGGGCGA | 实时荧光定量PCR RT-qPCR |
| GhNFD4-R | CTTTCGGGTACTTCCCTCCG | |
| GhNFD4-EcoRIF | CGGAATTCGACAAAGAGGGCCAAAGATGC | 基因克隆 Gene cloning |
| GhNFD4-BamHIR | CGGGATCCTCACCCCACTCTCCATTTTGT |
Table 1 Primers used in this study
引物名称 Primer name | 引物序列 Primer sequence (5′-3′) | 用途Purpose |
|---|---|---|
| GhUBQ7-F | AGAGGTCGAGTCTTGGGACA | 内参基因Reference gene |
| GhUBQ7-R | GCTTGATCTTCTTGGGCTTG | |
| GhNFD4-F | AAACCTTGTCACTTGGGCGA | 实时荧光定量PCR RT-qPCR |
| GhNFD4-R | CTTTCGGGTACTTCCCTCCG | |
| GhNFD4-EcoRIF | CGGAATTCGACAAAGAGGGCCAAAGATGC | 基因克隆 Gene cloning |
| GhNFD4-BamHIR | CGGGATCCTCACCCCACTCTCCATTTTGT |
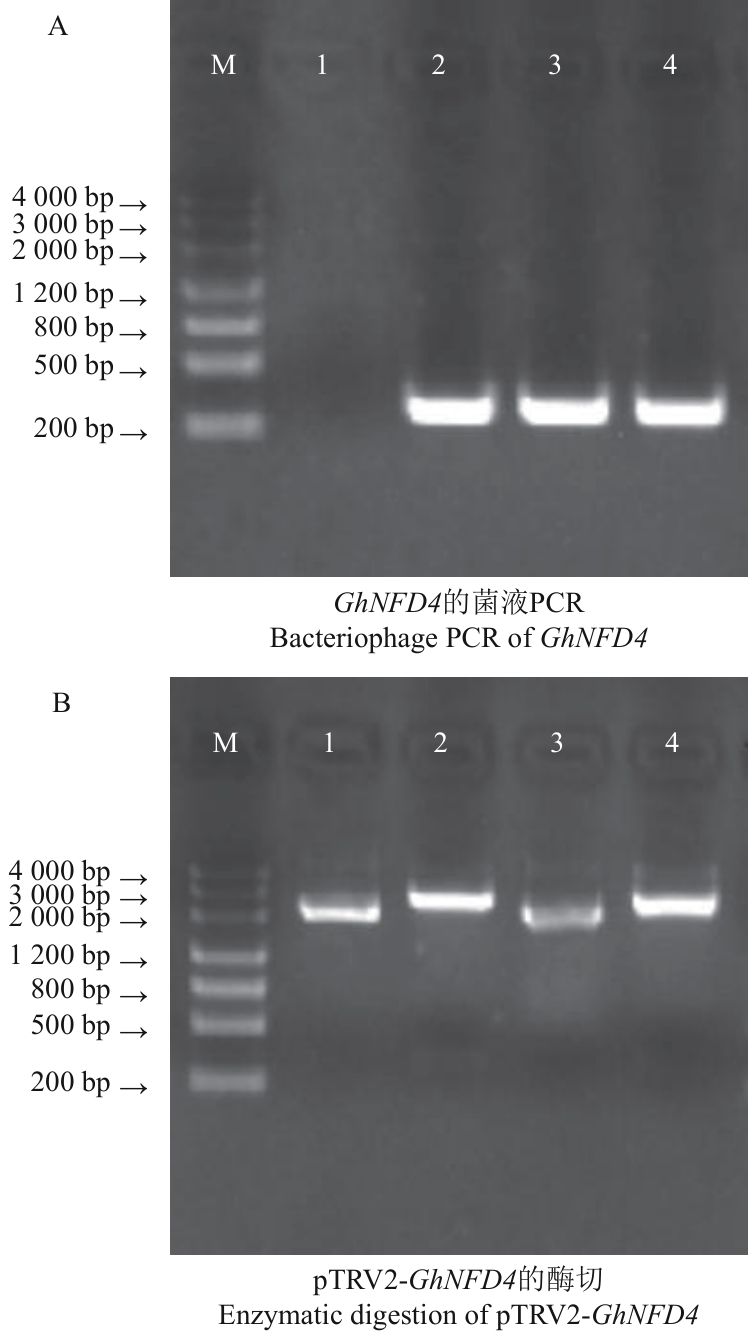
Fig. 2 Construction of GhNFD4 silencing vectorA: 1: Negative control; 2-4: cloning of GhNFD4 silencing fragment. B: 1-2: pTRV2-GhNFD4 (TRV2-NFD4-1) control and digest; 3-4: pTRV2-GhNFD4 (TRV2-NFD4-3) control and digest. M: Marker 3000
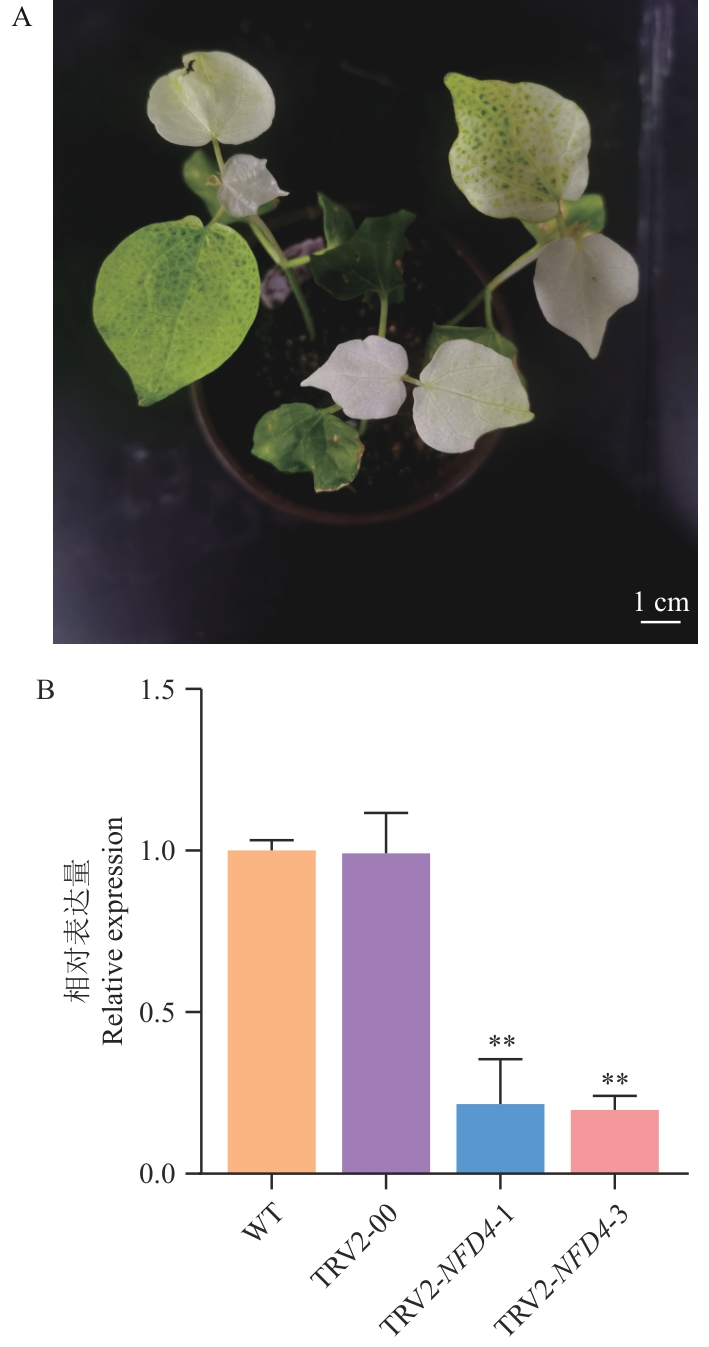
Fig. 3 Establishment of VIGS silencing system and detection of VIGS silencing efficiencyA: Cotton albino phenotype injected with pTRV1/pTRV2-GhCLA1. B: RT-qPCR analysis of GhNFD4 in WT, TRV2-00 and VIGS plants (TRV2-GhNFD4-1, TRV2-GhNFD4-3).**P<0.01. The same below
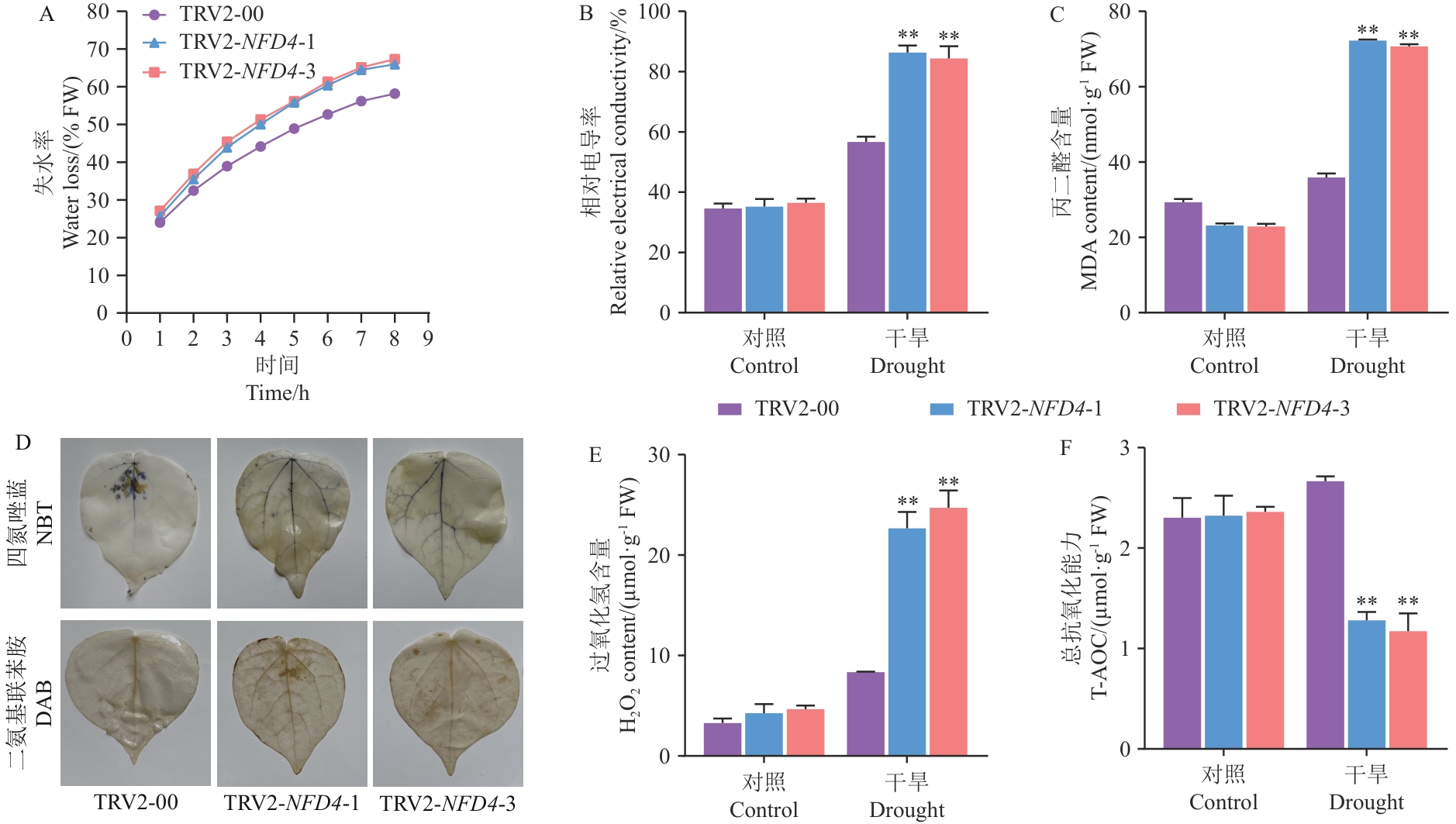
Fig. 5 Determination for the physiological indices of GhNFD4 gene-silenced plants measured after 15 d of drought stressA: Water loss rate of isolated leaves. B: Relative electrical conductivity. C: Malondialdehyde content (MDA). D: DAB and NBT staining of VIGS and control plants after drought stress. E: Hydrogen peroxide content (H2O2). F: Total antioxidant capacity (T-AOC)
| 1 | Bashir K, Matsui A, Rasheed S, et al. Recent advances in the characterization of plant transcriptomes in response to drought, salinity, heat, and cold stress [J]. F1000Res, 2019, 8(F1000 Faculty Rev): 658. |
| 2 | Mahmood U, Li XD, Fan YH, et al. Multi-omics revolution to promote plant breeding efficiency [J]. Front Plant Sci, 2022, 13: 1062952. |
| 3 | Abdelraheem A, Esmaeili N, O’Connell M, et al. Progress and perspective on drought and salt stress tolerance in cotton [J]. Ind Crops Prod, 2019, 130: 118-129. |
| 4 | Ergashovich KA, Azamatovna BZ, Toshtemirovna NU, et al. Ecophysiological effects of water deficiency on cotton varieties [J]. J Crit Rev, 2020, 7(9): 244-246. |
| 5 | Sajid M, Amjid M, Munir H, et al. Comparative analysis of growth and physiological responses of sugarcane elite genotypes to water stress and sandy loam soils [J]. Plants (Basel), 2023, 12(15): 2759. |
| 6 | Fang YJ, Liao KF, Du H, et al. A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice [J]. J Exp Bot, 2015, 66(21): 6803-6817. |
| 7 | Deeba F, Pandey AK, Ranjan S, et al. Physiological and proteomic responses of cotton (Gossypium herbaceum L.) to drought stress [J]. Plant Physiol Biochem, 2012, 53: 6-18. |
| 8 | Hussain S, Hussain S, Qadir T, et al. Drought stress in plants: an overview on implications, tolerance mechanisms and agronomic mitigation strategies [J]. Plant Sci Today, 2019, 6(4): 389-402. |
| 9 | Ozturk M, Turkyilmaz Unal B, García-Caparrós P, et al. Osmoregulation and its actions during the drought stress in plants [J]. Physiol Plant, 2021, 172(2): 1321-1335. |
| 10 | Pao SS, Paulsen IT, Jr Saier MH. Major facilitator superfamily [J]. Microbiol Mol Biol Rev, 1998, 62(1): 1-34. |
| 11 | Quistgaard EM, Löw C, Guettou F, et al. Understanding transport by the major facilitator superfamily (MFS): structures pave the way [J]. Nat Rev Mol Cell Biol, 2016, 17(2): 123-132. |
| 12 | Hwang JU, Song WY, Hong D, et al. Plant ABC transporters enable many unique aspects of a terrestrial plant’s lifestyle [J]. Mol Plant, 2016, 9(3): 338-355. |
| 13 | Guo RZ, Zhang Q, Qian K, et al. Phosphate-dependent regulation of vacuolar trafficking of OsSPX-MFSs is critical for maintaining intracellular phosphate homeostasis in rice [J]. Mol Plant, 2023, 16(8): 1304-1320. |
| 14 | Julião MHM, Silva SR, Ferro JA, et al. A genomic and transcriptomic overview of mate, abc, and MFS transporters in Citrus sinensis interaction with Xanthomonas citri subsp. citri [J]. Plants (Basel), 2020, 9(6): 794. |
| 15 | Boorer KJ, Loo DD, Wright EM. Steady-state and presteady-state kinetics of the H+/hexose cotransporter (STP1) from Arabidopsis thaliana expressed in Xenopus oocytes [J]. J Biol Chem, 1994, 269(32): 20417-20424. |
| 16 | Cordoba E, Aceves-Zamudio DL, Hernández-Bernal AF, et al. Sugar regulation of sugar transporter protein 1 (stp1) expression in Arabidopsis thaliana [J]. J Exp Bot, 2015, 66(1): 147-159. |
| 17 | Büttner M, Truernit E, Baier K, et al. AtSTP3, a green leaf-specific, low affinity monosaccharide-H+ symporter of Arabidopsis thaliana [J]. Plant Cell Environ, 2000, 23(2): 175-184. |
| 18 | Segonzac C, Boyer JC, Ipotesi E, et al. Nitrate efflux at the root plasma membrane: identification of an Arabidopsis excretion transporter [J]. Plant Cell, 2007, 19(11): 3760-3777. |
| 19 | Liu WW, Sun Q, Wang K, et al. Nitrogen Limitation Adaptation (NLA) is involved in source-to-sink remobilization of nitrate by mediating the degradation of NRT1.7 in Arabidopsis [J]. New Phytol, 2017, 214(2): 734-744. |
| 20 | Taochy C, Gaillard I, Ipotesi E, et al. The Arabidopsis root stele transporter NPF2.3 contributes to nitrate translocation to shoots under salt stress [J]. Plant J, 2015, 83(3): 466-479. |
| 21 | Liu TY, Huang TK, Yang SY, et al. Identification of plant vacuolar transporters mediating phosphate storage [J]. Nat Commun, 2016, 7: 11095. |
| 22 | Lu JY, Wang CL, Wang HY, et al. OsMFS1/OsHOP2 complex participates in rice male and female development [J]. Front Plant Sci, 2020, 11: 518. |
| 23 | Kong H, Hou MJ, Ma B, et al. Calcium-dependent protein kinase GhCDPK4 plays a role in drought and abscisic acid stress responses [J]. Plant Sci, 2023, 332: 111704. |
| 24 | Mandel MA, Feldmann KA, Herrera-Estrella L, et al. CLA1 a novel gene required for chloroplast development, is highly conserved in evolution [J]. Plant J, 1996, 9(5): 649-658. |
| 25 | Wei TL, Wang Y, Xie ZZ, et al. Enhanced ROS scavenging and sugar accumulation contribute to drought tolerance of naturally occurring autotetraploids in Poncirus trifoliata [J]. Plant Biotechnol J, 2019, 17(7): 1394-1407. |
| 26 | Zou LP, Qi DF, Sun JB, et al. Expression of the cassava nitrate transporter NRT2.1 enables Arabidopsis low nitrate tolerance [J]. J Genet, 2019, 98: 74. |
| 27 | Yan HL, Xu WX, Xie JY, et al. Variation of a major facilitator superfamily gene contributes to differential cadmium accumulation between rice subspecies [J]. Nat Commun, 2019, 10(1): 2562. |
| 28 | Zhang XM, Feng JJ, Zhao RL, et al. Functional characterization of the GhNRT2.1e gene reveals its significant role in improving nitrogen use efficiency in Gossypium hirsutum [J]. PeerJ, 2023, 11: e15152. |
| 29 | Liu FJ, Cai S, Dai LJ, et al. Two PHOSPHATE-TRANSPORTER1 genes in cotton enhance tolerance to phosphorus starvation [J]. Plant Physiol Biochem, 2023, 204: 108128. |
| 30 | Qiao KK, Lv JY, Chen LL, et al. GhSTP18, a member of sugar transport proteins family, negatively regulates salt stress in cotton [J]. Physiol Plant, 2023, 175(4): e13982. |
| 31 | Chen BZ, Wang XY, Lv JY, et al. GhN/AINV13 positively regulates cotton stress tolerance by interacting with the 14-3-3 protein [J]. Genomics, 2021, 113(1 Pt 1): 44-56. |
| 32 | Zhang Q, Ma C, Wang X, et al. Genome-wide identification of the light-harvesting chlorophyll a/b binding (Lhc) family in Gossypium hirsutum reveals the influence of GhLhcb2.3 on chlorophyll a synthesis [J]. Plant Biol, 2021, 23(5): 831-842. |
| 33 | Hano C, Drouet S, Lainé E. Virus-induced gene silencing (VIGS) in flax (Linum usitatissimum L.) seed coat: description of an effective procedure using the transparent testa 2 gene as a selectable marker [J]. Methods Mol Biol, 2020, 2172: 233-242. |
| [1] | LIU Jie, WANG Fei, TAO Ting, ZHANG Yu-jing, CHEN Hao-ting, ZHANG Rui-xing, SHI Yu, ZHANG Yi. Overexpression of SlWRKY41 Improves the Tolerance of Tomato Seedlings to Drought [J]. Biotechnology Bulletin, 2025, 41(2): 107-118. |
| [2] | HUANG Ying, YU Wen-jing, LIU Xue-feng, DIAO Gui-ping. Bioinformatics and Expression Pattern Analysis of Glutathione S-transferase in Populus davidiana × P. bolleana [J]. Biotechnology Bulletin, 2025, 41(2): 248-256. |
| [3] | DING Ruo-xi, DOU Shuo, AN Ye-zhi, KONG Wen-hui, GUO Wen-jing, ZHANG Dong-mei, WANG Xing-fen, MA Zhi-ying, WU Li-zhu. Extended Research on the Application of VIGS Technology in the Whole Growth Cycle of Cotton [J]. Biotechnology Bulletin, 2025, 41(2): 58-64. |
| [4] | KONG Qing-yang, ZHANG Xiao-long, LI Na, ZHANG Chen-jie, ZHANG Xue-yun, YU Chao, ZHANG Qi-xiang, LUO Le. Identification and Expression Analysis of GRAS Transcription Factor Family in Rosa persica [J]. Biotechnology Bulletin, 2025, 41(1): 210-220. |
| [5] | ZHANG Man-yu, DONG Jia-cheng, GOU Fu-fan, GONG Chao-hui, LIU Qian, SUN Wen-liang, KONG zhen, HAO Jie, WANG Min, TIAN Chao-guang. Cloning, Expression, Characterization and Application of the Pectin Esterase MtCE12-1 from Myceliophthora thermophila [J]. Biotechnology Bulletin, 2024, 40(9): 291-300. |
| [6] | WANG Qian, ZHOU Jia-yan, WANG Qian, DENG Yu-ping, ZHANG Min-hui, CHEN Jing, YANG Jun, ZOU Jian. Identification and Expression Analysis of the YABBY Gene Family in Sunflower [J]. Biotechnology Bulletin, 2024, 40(8): 199-211. |
| [7] | HAN Kai, ZHOU Yong-shun, ZHANG Kai-yue, WANG Lu, GAO Jian-feng, CHEN Fu-long. Evaluation of Drought Resistance of Three Chlorella Strains [J]. Biotechnology Bulletin, 2024, 40(8): 244-254. |
| [8] | YANG Wei, ZHAO Li-fen, TANG Bing, ZHOU Lin-bi, YANG Juan, MO Chuan-yuan, ZHANG Bao-hui, LI Fei, RUAN Song-lin, DENG Ying. Genome-wide Identification and Expression Analysis of the SRO Gene Family in Brassica juncea L. [J]. Biotechnology Bulletin, 2024, 40(8): 129-141. |
| [9] | HU Ya-dan, WU Guo-qiang, LIU Chen, WEI Ming. Roles of MYB Transcription Factor in Regulating the Responses of Plants to Stress [J]. Biotechnology Bulletin, 2024, 40(6): 5-22. |
| [10] | QIN Jian, LI Zhen-yue, HE Lang, LI Jun-ling, ZHANG Hao, DU Rong. Change of Single-cell Transcription Profile and Analysis of Intercellular Communication in Myogenic Cell Differentiation [J]. Biotechnology Bulletin, 2024, 40(6): 330-342. |
| [11] | WEN Jie, DU Yuan-xin, WU An-bo, YANG Guang-rong, LU Min, AN Hua-ming, NAN Hong. Identification and Expression Pattern Analysis of Rosa roxburghii SOD Gene Family [J]. Biotechnology Bulletin, 2024, 40(5): 153-166. |
| [12] | CHEN Chun-lin, LI Bai-xue, LI Jin-ling, DU Qing-jie, LI Meng, XIAO Huai-juan. Identification and Expression Analysis of Epidermal Patterning Factor (EPF) Genes in Cucumis melo [J]. Biotechnology Bulletin, 2024, 40(4): 130-138. |
| [13] | CHEN Qiang, HUANG Xin-hui, ZHANG Zheng, ZHANG Chong, LIU Ye-fei. Effects of Melatonin on the Fruit Softening and Ethylene Synthesis of Post-harvest Oriental Melon [J]. Biotechnology Bulletin, 2024, 40(4): 139-147. |
| [14] | YANG Wei-cheng, SUN Yan, YANG Qian, WANG Zhuang-lin, MA Ju-hua, XUE Jin-ai, LI Run-zhi. Genome-wide Identification of the FAX family in Gossypium hirsutum and Functional Analysis of GhFAX1 [J]. Biotechnology Bulletin, 2024, 40(3): 155-169. |
| [15] | ZHANG Yu, SHI Lei, GONG Lei, NIE Feng-jie, YANG Jiang-wei, LIU Xuan, YANG Wen-jing, ZHANG Guo-hui, XIE Rui-xia, ZHANG Li. Genome-wide Identification of Potato WOX Gene Family and Its Expression Analysis in in vitro Regeneration and Abiotic Stress [J]. Biotechnology Bulletin, 2024, 40(3): 170-180. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||