








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (11): 134-142.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0223
YE Yan( ), WU Yu-xuan, ZHOU Zhe-min, CUI Wen-jing(
), WU Yu-xuan, ZHOU Zhe-min, CUI Wen-jing( )
)
Received:2025-03-04
Online:2025-11-26
Published:2025-12-09
Contact:
CUI Wen-jing
E-mail:1815143307@qq.com;wjcui@jiangnan.edu.cn
YE Yan, WU Yu-xuan, ZHOU Zhe-min, CUI Wen-jing. Exploration, Characterization, and Application of Transaminase New Enzymes in the Biocatalytic Conversion of 2-aminobutyric Acid[J]. Biotechnology Bulletin, 2025, 41(11): 134-142.
菌株与质粒 Strain and plasmid | 性质 Properties | 来源 Source |
|---|---|---|
| E.coli BL21(DE3) | 表达宿主 | 实验室保存 |
| E.coli JM109 | 克隆宿主 | 实验室保存 |
| pET-28a-EsRTA | His 标签、Kan 抗性、T7 启动子 | 本实验构建 |
| pET-28a-Ec4a | His 标签、Kan 抗性、T7 启动子 | 本实验构建 |
| pET-28a-Bs | His 标签、Kan 抗性、T7 启动子 | 本实验构建 |
| pET-28a-CbRTA | His 标签、Kan 抗性、T7 启动子 | 本实验构建 |
| pET-28a-AtRTA | His 标签、Kan 抗性、T7 启动子 | 本实验构建 |
| pET-28a-TsRTA | His 标签、Kan 抗性、T7 启动子 | 本实验构建 |
| pET-28a-TsRTA | His 标签、Kan 抗性、T7 启动子 | 本实验构建 |
| Pbad-Bsalss | AmpR 抗性、araBAD 启动子 | 本实验构建 |
Table 1 Main strains and plasmids used in this study
菌株与质粒 Strain and plasmid | 性质 Properties | 来源 Source |
|---|---|---|
| E.coli BL21(DE3) | 表达宿主 | 实验室保存 |
| E.coli JM109 | 克隆宿主 | 实验室保存 |
| pET-28a-EsRTA | His 标签、Kan 抗性、T7 启动子 | 本实验构建 |
| pET-28a-Ec4a | His 标签、Kan 抗性、T7 启动子 | 本实验构建 |
| pET-28a-Bs | His 标签、Kan 抗性、T7 启动子 | 本实验构建 |
| pET-28a-CbRTA | His 标签、Kan 抗性、T7 启动子 | 本实验构建 |
| pET-28a-AtRTA | His 标签、Kan 抗性、T7 启动子 | 本实验构建 |
| pET-28a-TsRTA | His 标签、Kan 抗性、T7 启动子 | 本实验构建 |
| pET-28a-TsRTA | His 标签、Kan 抗性、T7 启动子 | 本实验构建 |
| Pbad-Bsalss | AmpR 抗性、araBAD 启动子 | 本实验构建 |

Fig. 1 Structural clustering and sequence homology analysis of transaminases and recombinant expression of SDS-PAGEA: Structural clustering; B: sequence homology analysis; C: recombinant expression of SDS-PAGE (b indicates bacterial liquid, s indicates supernatant of lysed bacterial liquid)

Fig. 3 Optimal temperature, optimal pH, temperature stability and pH stability of Ec4aA: Optimal temperature for Ec4a. B: Optimal pH for Ec4a. C: Temperature stability of Ec4a. D: pH stability of Ec4a
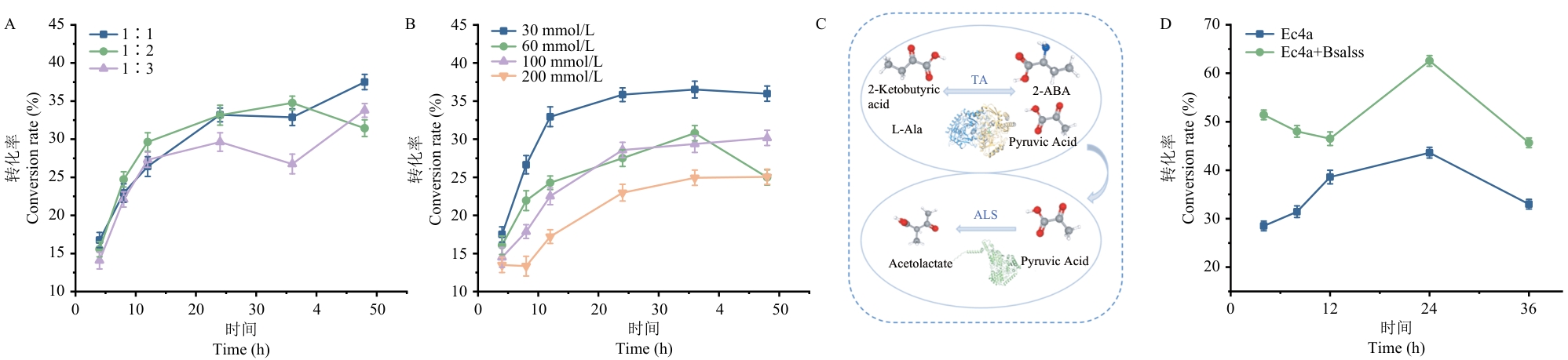
Fig. 5 Optimization of whole-cell catalytic systems and biocatalysis cascade systemsA: Conversion rates under different substrate ratios. B: Conversion rates under different substrate concentrations. C: Schematic diagram of the cascading system. D: Catalytic conversion rate of the control group
| [1] | Kelly SA, Pohle S, Wharry S, et al. Application of ω-transaminases in the pharmaceutical industry [J]. Chem Rev, 2018, 118(1): 349-367. |
| [2] | 蔡雪, 孙晨阳, 翟增春, 等. 磷酸吡哆醛依赖型酶的研究进展及其应用[J]. 生物工程学报, 2024, 40(9): 2771-2785. |
| Cai X, Sun CY, Zhai ZC, et al. Research progress and applications of pyridoxal phosphate-dependent enzymes [J]. Chinese Journal of Biotechnology, 2024, 40(9): 2771-2785. | |
| [3] | Slabu I, Galman JL, Lloyd RC, et al. Discovery, engineering, and synthetic application of transaminase biocatalysts [J]. ACS Catalysis, 2017, 7(12): 8263-8284. |
| [4] | Farkas E, Sátorhelyi P, Szakács Z, et al. Transaminase-catalysis to produce trans-4-substituted cyclohexane-1-amines including a key intermediate towards cariprazine [J]. Commun Chem, 2024, 7(1): 86. |
| [5] | 夏温娜, 孙雨, 闵聪, 等. 转氨酶催化不对称合成芳香族L-氨基酸 [J]. 生物工程学报, 2012, 28(11): 1346-1358. |
| Xia WN, Sun Y, Min C, et al. Asymmetric synthesis of aromatic L-amino acids catalyzed by transaminase [J]. Chin J Biotechnol, 2012, 28(11): 1346-1358. | |
| [6] | Park ES, Dong JY, Shin JS. ω-Transaminase-catalyzed asymmetric synthesis of unnatural amino acids using isopropylamine as an amino donor [J]. Org Biomol Chem, 2013, 11(40): 6929-6933. |
| [7] | Babu KC, Reddy RB, Mukkanti K, et al. Enantioselective synthesis of antiepileptic agent, (-)-levetiracetam, through Evans Asymmetric strategy [J]. Journal of Chemistry, 2013, 72(64): 13846-13855. |
| [8] | Chen JJ, Zhu RS, Zhou JP, et al. Efficient single whole-cell biotransformation for L-2-aminobutyric acid production through engineering of leucine dehydrogenase combined with expression regulation [J]. Bioresour Technol, 2021, 326: 124665. |
| [9] | 张利坤, 肖延铭, 杨卫华, 等. 亮氨酸脱氢酶偶联NADH再生体系合成L-2-氨基丁酸 [J]. 生物工程学报, 2020, 36(5): 992-1001. |
| Zhang LK, Xiao YM, Yang WH, et al. Synthesis of L-2-aminobutyric acid by leucine dehydrogenase coupling with an NADH regeneration system [J]. Chin J Biotechnol, 2020, 36(5): 992-1001. | |
| [10] | 奚强, 丁友友, 林丫丫, 等. 2-氨基丁酸的酶拆分[J]. 武汉工程大学学报, 2009, 31(3): 26-29. |
| Xi Q, Ding YY, Lin YY, et al. Enzymatic resolution of 2-aminobutyric acid [J]. Journal of Wuhan Institute of Technology, 2009, 31(3): 26-29. | |
| [11] | Liu YF, Han LC, Cheng ZY, et al. Enzymatic biosynthesis of L-2-aminobutyric acid by glutamate mutase coupled with L-Aspartate-β-decarboxylase using L-glutamate as the sole substrate [J]. ACS Catalysis, 2020, 10(23): 13913-13917. |
| [12] | Shin JS, Kim BG. Transaminase-catalyzed asymmetric synthesis of L-2-aminobutyric acid from achiral reactants [J]. Biotechnol Lett, 2009, 31(10): 1595-1599. |
| [13] | Tao RS, Jiang Y, Zhu FY, et al. A one-pot system for production of L-2-aminobutyric acid from L-threonine by L-threonine deaminase and a NADH-regeneration system based on L-leucine dehydrogenase and formate dehydrogenase [J]. Biotechnol Lett, 2014, 36(4): 835-841. |
| [14] | 周俊平. L-2-氨基丁酸的微生物高效制备及关键酶的理性改造 [D]. 无锡:江南大学, 2019. |
| Zhou JP, Microbial high-efficiency preparation of L-2-aminobutyric acid and rational modification of key enzymes [D]. Wuxi: Jiangnan University, 2019. | |
| [15] | Zhang ZW, Liu Y, Zhao J, et al. Active-site engineering of ω- transaminase from Ochrobactrum anthropi for preparation of L-2-aminobutyric acid [J]. BMC Biotechnol, 2021, 21(1): 55. |
| [16] | Luo W, Hu JG, Lu JP, et al. One pot cascade synthesis of L-2-aminobutyric acid employing ω-transaminase from Paracoccus pantotrophus [J]. Mol Catal, 2021, 515: 111890. |
| [17] | Cui YM, Gao YD, Yang LC. Transaminase catalyzed asymmetric synthesis of active pharmaceutical ingredients [J]. Green Synth Catal, 2024. |
| [18] | Gomm A, O'Reilly E. Transaminases for chiral amine synthesis [J]. Curr Opin Chem Biol, 2018, 43: 106-112. |
| [19] | Pinto A, Contente ML, Tamborini L. Advances on whole-cell biocatalysis in flow [J]. Curr Opin Green Sustain Chem, 2020, 25: 100343. |
| [20] | Lin BX, Tao Y. Whole-cell biocatalysts by design [J]. Microb Cell Fact, 2017, 16(1): 106. |
| [21] | 胡佳桂. ω-转氨酶基因挖掘、级联反应体系构建及其应用 [D]. 无锡: 江南大学, 2021. |
| Hu JG. Gene mining of ω-aminotransferase, construction of cascade reaction system and its application [D]. Wuxi: Jiangnan University, 2021. | |
| [22] | Jiang JJ, Chen X, Zhang DL, et al. Characterization of (R)-selective amine transaminases identified by in silico motif sequence blast [J]. Appl Microbiol Biotechnol, 2015, 99(6): 2613-2621. |
| [23] | Kelly SA, Mix S, Moody TS, et al. Transaminases for industrial biocatalysis: novel enzyme discovery [J]. Appl Microbiol Biotechnol, 2020, 104(11): 4781-4794. |
| [24] | 蒙丽钧. 转氨酶在大肠杆菌中的克隆表达及应用 [D]. 杭州: 浙江大学, 2019. |
| Meng LJ. Cloning and expression of transaminase in Escherichia coli and its application [D]. Hangzhou: Zhejiang University, 2019. | |
| [25] | 蔡婷婷, 曹佳仁, 邱帅, 等. 半理性设计进化土曲霉来源的ω-转氨酶AtTA热稳定性 [J]. 生物工程学报, 2023, 39(6): 2126-2140. |
| Cai TT, Cao JR, Qiu S, et al. Semi-rational evolution of ω-transaminase from Aspergillus terreus for enhancing the thermostability [J]. Chin J Biotechnol, 2023, 39(6): 2126-2140. | |
| [26] | Meng QL, Ramírez-Palacios C, Wijma HJ, et al. Protein engineering of amine transaminases [J]. Frontiers in Catalysis, 2022, 2. DOI: 10.3389/fctls.2022.1049179 |
| [27] | Khatik AG, Muley AB, More PR, et al. Transaminase-mediated chiral selective synthesis of (1R)-(3-methylphenyl)ethan-1-amine from 1-(3-methylphenyl)ethan-1-one: process minutiae, optimization, characterization and ‘What If studies’ [J]. Bioprocess Biosyst Eng, 2023, 46(2): 207-225. |
| [28] | 郑裕国, 程峰, 金利群, 等. 一种转氨酶突变体及其生产L-草铵膦的应用:中国,CN201810540686.1 [P]. 2018-05-30. |
| Zheng YG, Cheng F, Jin LQ, et al. A transaminase mutant and its application in the production of L-phosphinothricin: China, CN201810540686.1 [P]. 2018-05-30. | |
| [29] | 李仲霞, 刘妍, 罗泉, 等. 从专利角度分析ω-转氨酶在我国手性胺生物合成应用中的研究进展 [J]. 生物工程学报, 2023, 39(8): 3169-3187. |
| Li ZX, Liu Y, Luo Q, et al. The advance of ω-transaminase in chiral amine biosynthesis in China from the perspective of patents [J]. Chin J Biotechnol, 2023, 39(8): 3169-3187. | |
| [30] | Cárdenas-Fernández M, Sinclair O, Ward JM. Novel transaminases from thermophiles: from discovery to application [J]. Microb Biotechnol, 2022, 15(1): 305-317. |
| [31] | Wang ZC, Xu ML, Xie YY, et al. One-pot two-stage biocatalytic cascade to produce l-phosphinothricin by two enantioselective complementary aminotransferases at high substrate loading via a deracemization process [J]. J Agric Food Chem, 2024. |
| [32] | Höhne M, Kühl S, Robins K, et al. Efficient asymmetric synthesis of chiral amines by combining transaminase and pyruvate decarboxylase [J]. Chembiochem, 2008, 9(3): 363-365. |
| [33] | Guo F, Berglund P. Transaminase biocatalysis: optimization and application [J]. Green Chem, 2017, 19(2): 333-360. |
| [1] | WANG Hao, CAO An-ni, GAO Xin-yi, GUO Min-liang. Enzymatic Characterization and Directed Evolution of Agrobacterium tumefaciens O-demethylase Atu1420 [J]. Biotechnology Bulletin, 2025, 41(3): 319-329. |
| [2] | WANG Bi-cheng, JING Hai-qing, WAN Kun, ZHANG Ying-ying, DING Jia-hao, LI Run-zhi, XUE Jin-ai, ZHANG Hai-ping. Identification of Soybean BCAT Gene Family and Functional Analysis of GmBCAT3 in Soybean Responses to Drought Stress [J]. Biotechnology Bulletin, 2025, 41(10): 196-209. |
| [3] | ZHANG Man-yu, DONG Jia-cheng, GOU Fu-fan, GONG Chao-hui, LIU Qian, SUN Wen-liang, KONG zhen, HAO Jie, WANG Min, TIAN Chao-guang. Cloning, Expression, Characterization and Application of the Pectin Esterase MtCE12-1 from Myceliophthora thermophila [J]. Biotechnology Bulletin, 2024, 40(9): 291-300. |
| [4] | ZHENG Fei, YANG Jun-zhao, NIU Yu-feng, LI Rui-lin, ZHAO Guo-zhu. Characterization and Functional Analysis of Lytic Polysaccharide Monooxygenase TtLPMO9I from Thermothelomyces thermophilus [J]. Biotechnology Bulletin, 2024, 40(2): 289-299. |
| [5] | QU Ge, SUN Zhou-tong. Catalytic Promiscuity-driven Redesign of Enzyme Functions [J]. Biotechnology Bulletin, 2023, 39(4): 1-9. |
| [6] | ZHAO Sai-sai, ZHANG Xiao-dan, JIA Xiao-yan, TAO Da-wei, LIU Ke-yu, NING Xi-bin. Investigation on the Complex Mutagenesis Selection of High-yield Nitrate Reductase Strain Staphylococcus simulans ZSJ6 and Its Enzymatic Properties [J]. Biotechnology Bulletin, 2023, 39(4): 103-113. |
| [7] | WANG Mu-qiang, CHEN Qi, MA Wei, LI Chun-xiu, OUYANG Peng-fei, XU Jian-he. Advances in the Application of Machine Learning Methods for Directed Evolution of Enzymes [J]. Biotechnology Bulletin, 2023, 39(4): 38-48. |
| [8] | ZHANG Kai-ping, LIU Yan-li, TU Mian-liang, LI Ji-wei, WU Wen-biao. Optimization of Producing Cellulase by Aspergillus fumigatus A-16 and Its Enzymatic Properties [J]. Biotechnology Bulletin, 2022, 38(9): 215-225. |
| [9] | CHANG Qing, SHU Yue-rong, WANG Wen-tao, JIANG Hao, YAN Quan-de, QIAN Zheng, GAO Xue-chun, WU Jin-hong, ZHANG Yong. Heterologous Expression and Characterization of Endo-type Alginate Lyase from Yeosuana marina sp. JLT21 [J]. Biotechnology Bulletin, 2022, 38(2): 123-131. |
| [10] | TIAN Jia-hui, FENG Jia-li, LU Jun-hua, MAO Lin-jing, HU Zhu-ran, WANG Ying, CHU Jie. Isolation,Purification and Characterization of Laccase LacT-1 from Cerrena unicolor [J]. Biotechnology Bulletin, 2021, 37(8): 186-194. |
| [11] | BAI Fu-mei, LI Zhi-min, WANG Xiao-qin, HU Zi-wei, BAO Ling-ling, LI Zhi-min. Biochemical Characterization and Structural Analysis of N-acetylornithine Transaminase from Synechocystis sp. PCC6803 [J]. Biotechnology Bulletin, 2021, 37(5): 98-107. |
| [12] | LIU Shan, YE Wei, ZHU Mu-zi, LI Sai-ni, DENG Zhang-shuang, ZHANG Wei-min. Cloning,Expression and Characterization of a Novel Acyltransferase GPAT [J]. Biotechnology Bulletin, 2021, 37(11): 257-266. |
| [13] | ZHAO Hai-yan, SONG Chen-bin, LIU Zheng-ya, MA Xing-rong, SHANG Hui-hui, LI An-hua, GUAN Xian-jun, WANG Jian-she. Cloning,Recombinant Expression and Enzymatic Properties of α-Amylase Gene from Laceyella sp. [J]. Biotechnology Bulletin, 2020, 36(8): 23-33. |
| [14] | ZHU Cai-lin, LÜ Xiang, XIA Xiao-le. Effect of Site-directed Mutagenesis of Amino Acids in Lid Region on the Enzymatic Properties of T1 Lipase [J]. Biotechnology Bulletin, 2020, 36(11): 94-102. |
| [15] | GUO Jing-jing, GUO Lei-lei, ZHAO Yun-xiu, DAI Yi-jun. Research on the NAMase of Ensifer meliloti 1021 and Regulation Mechanism of 3-Cyanopyridine [J]. Biotechnology Bulletin, 2019, 35(8): 51-58. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||