








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (11): 121-133.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0372
YANG Yi-chen1( ), ZHU Hong-yu2, SU Xiao-yun2, WANG Yuan2, LUO Hui-ying2, TIAN Jian2, YAO Bin1,2, HUANG Huo-qing2(
), ZHU Hong-yu2, SU Xiao-yun2, WANG Yuan2, LUO Hui-ying2, TIAN Jian2, YAO Bin1,2, HUANG Huo-qing2( ), ZHANG Jie2(
), ZHANG Jie2( )
)
Received:2025-04-09
Online:2025-11-26
Published:2025-12-09
Contact:
HUANG Huo-qing, ZHANG Jie
E-mail:xms02yyc@163.com;huanghuoqing@caas.cn;zhangjie09@caas.cn
YANG Yi-chen, ZHU Hong-yu, SU Xiao-yun, WANG Yuan, LUO Hui-ying, TIAN Jian, YAO Bin, HUANG Huo-qing, ZHANG Jie. Construction of an Efficient Microbial Cell Factory for Inositol Production from Glucose-fructose Syrup[J]. Biotechnology Bulletin, 2025, 41(11): 121-133.
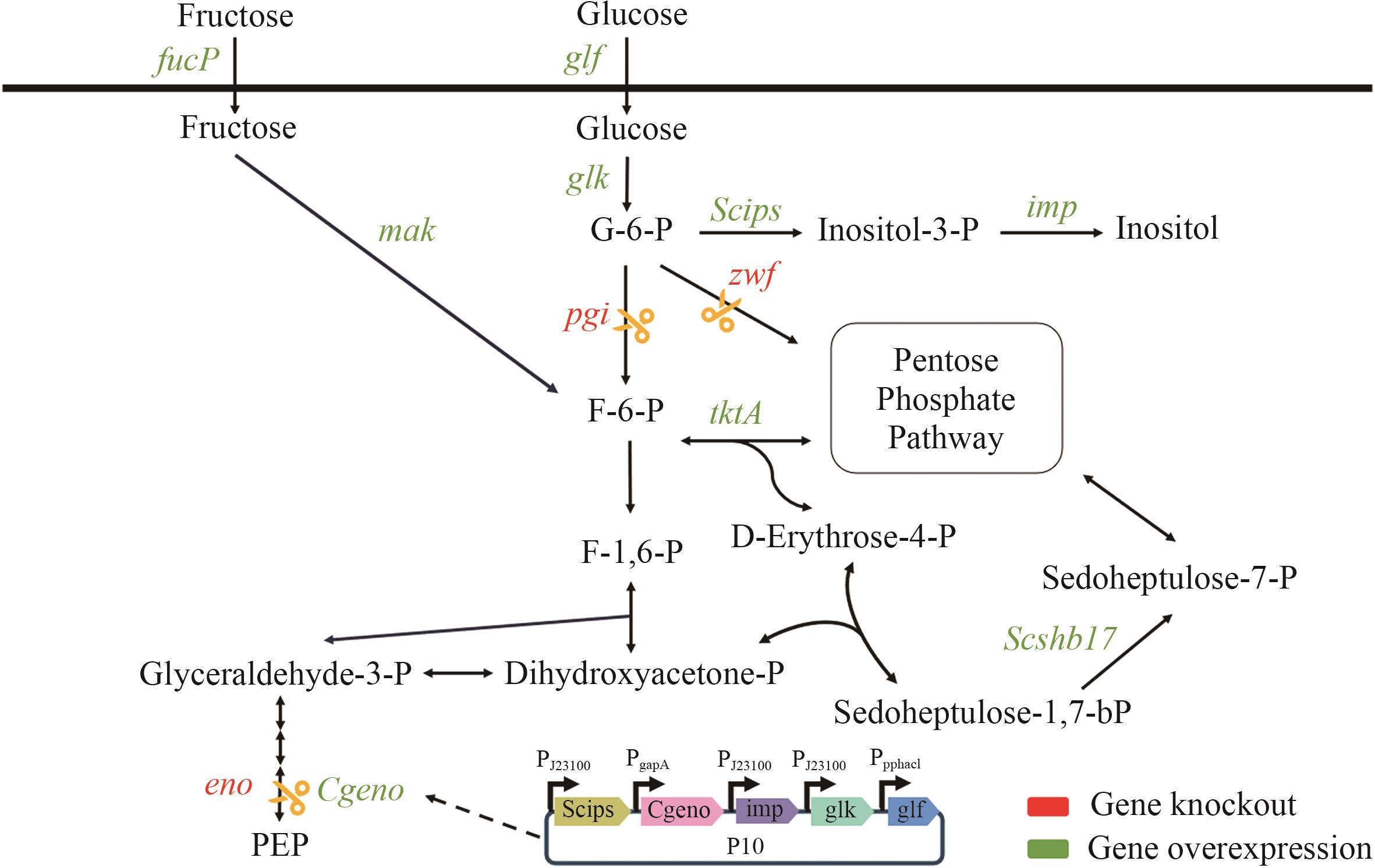
Fig. 1 Inositol biosynthesis pathway using fructose and glucose as substrateglf: Glucose facilitator gene. glk: Glucokinase gene. imp: Inositol monophosphatase gene. Scips: Inositol-3-phosphate synthase gene derived from Saccharomyces cerevisiae. pgi: Glucose-6-phosphate isomerase gene. zwf: Glucose-6-phosphate dehydrogenase gene. tktA: Transketolase gene. fucP: Fructose transporter gene. mak: Fructokinase gene. Scshb17: Sedoheptulose bisphosphatase gene. Cgeno: Enolase gene derived from Corynebacterium glutamicum
菌株/质粒 Strains/Plasmids | 相关特性 Relevant characteristics | 来源 Sources |
|---|---|---|
| Strains | ||
| E. coli Trans1-T1 | Commercial host for cloning | TransGen Biotech |
| E. coli BW25113 | Wild type, starting strain | Lab storage |
| JY1 | E. coli BW25113ΔpgiΔzwf | This study |
| JY2 | JY1 harboring plasmid P5 | This study |
| JY3 | JY1ΔgapC1::P gapA -Scshb17 | This study |
| JY4 | JY1ΔP tktA ::P gapA -Scshb17-P J23100 | This study |
| JY5 | JY1ΔS9::P gapA -fucP | This study |
| JY6 | JY3ΔS9::P gapA -fucP | This study |
| JY7 | JY4ΔS9::P gapA -fucP | This study |
| JY8 | JY4ΔP mak ::P gapA -fucP-P J23100 | This study |
| JY9 | JY6 harboring plasmid P5 | This study |
| JY10 | JY7 harboring plasmid P5 | This study |
| JY11 | JY6ΔLafu::P J23100 -imp; ΔydeU:: P J23100 -imp | This study |
| JY12 | JY11ΔycdN::P araBAD -Scips | This study |
| JY13 | JY11ΔycdN::P araBAD -Scips; ΔadhE::P araBAD -Scips | This study |
| JY14 | JY11ΔycdN::P araBAD -Scips; ΔadhE::P araBAD -Scips; ΔycjV::P araBAD -Scips | This study |
| JY15 | JY11ΔyciQ:: P J23100 -Scips | This study |
| JY16 | JY11ΔyciQ:: P J23100 -Scips; Δycgh:: P J23100 -Scips | This study |
| JY17 | JY6Δeno harboring plasmid P9 | This study |
| JY18 | JY6Δeno harboring plasmid P10 | This study |
| Plasmid | ||
| pTetQCas-BsaI | CloDF13 ori, SmR, Ptet-tniQ-cas876, Ptet-CRISPR (containing two BsaI restriction sites) | [ |
| pTet-tns | RSF1030 ori, KanR, Ptet-tnsABC | [ |
| pRE57-Ter | ColE1 ori, AmpR, Donor plasmid harboring Right end (57 bp)-terminator-Left end cassette | [ |
| pEASY-T3 | ColE1 ori, AmpR, TA cloning vector | Lab storage |
| pRSFDuet-1 | KanR, RSF ori, expression vector | Lab storage |
| pETDuet-1 | AmpR, ColE1 ori, expression vector | Lab storage |
| P1 | pEASY-T3 harboring sgRNA2-gapC; P gapA -Scshb17 | This study |
| P2 | pEASY-T3 harboring sgRNA-S9; P gapA -fucP | This study |
| P3 | pEASY-T3 harboring sgRNA-P tktA; P gapA -Scshb17-P J23100 | This study |
| P4 | pEASY-T3 harboring sgRNA-P mak; P gapA -fucp-P J23100 | This study |
| P5 | pRSFDuet-1 harboring P J23100 -Scips; P J23100 -imp | This study |
| P6 | pRE57-Ter harboring P J23100 -imp | This study |
| P7 | pRE57-Ter harboring P araBAD -Scips | This study |
| P8 | pRE57-Ter harboring P J23100 -Scips | This study |
| P9 | pRSFDuet-1 harboring P J23100 -Scips; P J23100 -imp; P gapA -Cgeno; CmR | This study |
| P10 | pRSFDuet-1 harboring P J23100 -Scips; P J23100 -imp; P gapA -Cgeno; P J23100 -glk; P phac1 -glf | This study |
Table 1 Strains and plasmids used in this work
菌株/质粒 Strains/Plasmids | 相关特性 Relevant characteristics | 来源 Sources |
|---|---|---|
| Strains | ||
| E. coli Trans1-T1 | Commercial host for cloning | TransGen Biotech |
| E. coli BW25113 | Wild type, starting strain | Lab storage |
| JY1 | E. coli BW25113ΔpgiΔzwf | This study |
| JY2 | JY1 harboring plasmid P5 | This study |
| JY3 | JY1ΔgapC1::P gapA -Scshb17 | This study |
| JY4 | JY1ΔP tktA ::P gapA -Scshb17-P J23100 | This study |
| JY5 | JY1ΔS9::P gapA -fucP | This study |
| JY6 | JY3ΔS9::P gapA -fucP | This study |
| JY7 | JY4ΔS9::P gapA -fucP | This study |
| JY8 | JY4ΔP mak ::P gapA -fucP-P J23100 | This study |
| JY9 | JY6 harboring plasmid P5 | This study |
| JY10 | JY7 harboring plasmid P5 | This study |
| JY11 | JY6ΔLafu::P J23100 -imp; ΔydeU:: P J23100 -imp | This study |
| JY12 | JY11ΔycdN::P araBAD -Scips | This study |
| JY13 | JY11ΔycdN::P araBAD -Scips; ΔadhE::P araBAD -Scips | This study |
| JY14 | JY11ΔycdN::P araBAD -Scips; ΔadhE::P araBAD -Scips; ΔycjV::P araBAD -Scips | This study |
| JY15 | JY11ΔyciQ:: P J23100 -Scips | This study |
| JY16 | JY11ΔyciQ:: P J23100 -Scips; Δycgh:: P J23100 -Scips | This study |
| JY17 | JY6Δeno harboring plasmid P9 | This study |
| JY18 | JY6Δeno harboring plasmid P10 | This study |
| Plasmid | ||
| pTetQCas-BsaI | CloDF13 ori, SmR, Ptet-tniQ-cas876, Ptet-CRISPR (containing two BsaI restriction sites) | [ |
| pTet-tns | RSF1030 ori, KanR, Ptet-tnsABC | [ |
| pRE57-Ter | ColE1 ori, AmpR, Donor plasmid harboring Right end (57 bp)-terminator-Left end cassette | [ |
| pEASY-T3 | ColE1 ori, AmpR, TA cloning vector | Lab storage |
| pRSFDuet-1 | KanR, RSF ori, expression vector | Lab storage |
| pETDuet-1 | AmpR, ColE1 ori, expression vector | Lab storage |
| P1 | pEASY-T3 harboring sgRNA2-gapC; P gapA -Scshb17 | This study |
| P2 | pEASY-T3 harboring sgRNA-S9; P gapA -fucP | This study |
| P3 | pEASY-T3 harboring sgRNA-P tktA; P gapA -Scshb17-P J23100 | This study |
| P4 | pEASY-T3 harboring sgRNA-P mak; P gapA -fucp-P J23100 | This study |
| P5 | pRSFDuet-1 harboring P J23100 -Scips; P J23100 -imp | This study |
| P6 | pRE57-Ter harboring P J23100 -imp | This study |
| P7 | pRE57-Ter harboring P araBAD -Scips | This study |
| P8 | pRE57-Ter harboring P J23100 -Scips | This study |
| P9 | pRSFDuet-1 harboring P J23100 -Scips; P J23100 -imp; P gapA -Cgeno; CmR | This study |
| P10 | pRSFDuet-1 harboring P J23100 -Scips; P J23100 -imp; P gapA -Cgeno; P J23100 -glk; P phac1 -glf | This study |
引物名称 Primer name | 引物序列 Primer sequence (5′-3′) |
|---|---|
| gapC2-up-F | CACTAGTGAATTCGCGGCCGCCTGCAGAACAGGAGATAAATCACCAAATGTCCCAAATGCGC |
| gapC2-up-R | AGAGCTCTCCCATATGGTCGACATAAGTGAATCTTCCGTAAAATCAACGC |
| gapC2-down-F | TCGACCATATGGGAGAGCTCTTTACGCCGAAAAAGAGGCTAAAAATATTC |
| gapC2-down-R | AATACTCAAGCTATGCATCCAACGCGTTGGGTTTGCGTGGCATCAAACACCGAACCG |
| gRNA2-gapC-F | TTGACAGCTAGCTCAGTCCTAGGTATAATGCTAGCCGTTGATGGGAAAAGTATCGGTTTTAGAGCTAGAAATAGCAAGTT |
| gRNA2-gapC-R | GGTTCTTATGGCTCTTGTATCTATCAGTGAAGCATCAAGAC |
| S9-up-F | CACTAGTGAATTCGCGGCCGCCTGCAGGCACGGAGAATGGCAGGAAGCATC |
| S9-up-R | CAATCCTACCGAGCTCTCCCATATGGTCGACCAGGTTTATTATATCGCGTTGATTATTGATGCTGTTTTTAGTTTTAACGGC |
| S9-down-F | TAATAAACCTGGTCGACCATATGGGAGAGCTCGGTAGGATTGAAAACGCTCTCCTGATTTTCCAATTCA |
| S9-down-R | GCTATGCATCCAACGCGTTGGGCAGCCCGCCCGGTTGCCCG |
| gRNA-S9-F | TTGACAGCTAGCTCAGTCCTAGGTATAATGCTAGCTCTGGCGCAGTTGATATGTAGTTTTAGAGCTAGAAATAGCAAGTT |
| gRNA-S9-R | GGTTCTTATGGCTCTTGTATCTATCAGTGAAGCATCAAGAC |
| gapA-Scshb17-F | ACGGAAGATTCACTTATGTCGACATCTCGACGAAATGGCTGCAC |
| gapA-Scshb17-R | CATCTGGGGGTTAGCGAAGGCATATATTCCACCAGCTATTTGTTAGTG |
| Scshb17-F | CACTAACAAATAGCTGGTGGAATATATGCCTTCGCTAACCCCCAG |
| Scshb17-R | TTAGCCTCTTTTTCGGCGTAAAGAGCTCTTACACATCGCCATGCTGGG |
| GapA-fucP-F | CAATAATCAACGCGATATAATAAACCTGATCTCGACGAAATGGCTGCAC |
| GapA-fucP-R | GTTTGTATTGATGTGTTTCCCATATATTCCACCAGCTATTTGTTAG |
| fucP-F | ACAAATAGCTGGTGGAATATATGGGAAACACATCAATACAAACGC |
| fucP-R | CAGGAGAGCGTTTTCAATCCTACCGAGCTCCATGGATCCTCAGTTAGTTGCCGTTTGAGAACG |
| gRNA1-gapC-F | TTGACAGCTAGCTCAGTCCTAGGTATAATGCTAGCCGATTTACAACTGGTGAAAAGTTTTAGAGCTAGAAATAGCAAGTT |
| gRNA1-gapC-R | GGTTCTTATGGCTCTTGTATCTATCAGTGAAGCATCAAGAC |
| gapC1-up-F | CACTAGTGAATTCGCGGCCGCCTGCAGCTAAAAATATTCCGTGGAAAGCGAAAGGTGCAGAAAT |
| gapC1-up-R | CCATATTCGTTATCGTACCAGGCGAGAGCTCTCCCATATGGTCGACCCCACGGCGGTAATTTCCGTTTGC |
| gapC1-down-F | CGGAAATTACCGCCGTGGGGTCGACCATATGGGAGAGCTCTCGCCTGGTACGATAACGAATATGGCTTCG |
| gapC1-down-R | CTCAAGCTATGCATCCAACGCGTTGGTTTATGATCAGTTCGTCAATCGGCTGGGTGAAGC |
| phac1+glf-F | GAGCTCTAGCTGGACGTCTAGAGTTGACAGCGCGTGCGTTGCAAG |
| Glf-R | GCCATATTCGTTATCGTACCAGGCGATTATTTCTGGCTGCGCCACATTTC |
| gRNA-P glk -F | TTGACAGCTAGCTCAGTCCTAGGTATAATGCTAGCTCTCACACTGTAAATACCTGGTTTTAGAGCTAGAAATAGCAAGTT |
| gRNA-P glk -R | GGTTCTTATGGCTCTTGTATCTATCAGTGAAGCATCAAGAC |
| glk-up-F | ATTCGCGGCCGCCTGCAGGTCGACCTATTCGGCGCAAAATCAACGT |
| glk-up-R | GAAAGAGGAGAAATACTAGATGACAAAGTATGCATTAGTCGGTGATGTGGGC |
| glk-down-F | CTTTGTCATCATATGACGGAGCTCAGGCAGGCTCCCTGTAAATATC |
| glk-down-R | CAAGCTATGCATCCAACGCGTTGGCGGTCATGATCGGATGTTCCG |
| gRNA-P tktA -F | TTGACAGCTAGCTCAGTCCTAGGTATAATGCTAGCGATCGGATGATGAAGGGCACGTTTTAGAGCTAGAAATAGCAAGTT |
| gRNA-P tktA -R | GGTTCTTATGGCTCTTGTATCTATCAGTGAAGCATCAAGAC |
| gapA-eno-F | CATTTGAGAAGCACACGGTCACACTGCATCTCGACGAAATGGCTGCAC |
| gapA-eno-R | ACGTGCATGATTTCAGCCATATATTCCACCAGCTATTTGTTAGTGAATAAAAG |
| Cgeno-F | CACTAACAAATAGCTGGTGGAATATATGGCTGAAATCATGCACGTATTC |
| Cgeno-R | GTGCGTCGGGTGATGCTGCCAACTTACTTAGCCCTGAAAGCGTGGGAATG |
| CmR-F | GTATCCGCTCATGAATTAATTCCAATAAACCGGTAAACCAGCAATAG |
| CmR-R | GCATTCCCACGCTTTCAGGGCTAAGTAAGTTGGCAGCATCACCCG |
| Scips-F | GAAAGAGGAGAAATACTAGATGACAGAAGATAATATTGCTCCAATCAC |
| imp-R | GACCGTGTGCTTCTCAAATGTTAACGCTTCAGAGCGTCGC |
| P tktA -gRNA | GATCGGATGATGAAGGGCAC |
| pgi-gRNA | TGGCAGGCACTACAGAAACACTTCGATGAAAT |
| zwf-gRNA | GAGACGGCTGAATGCAGCAGTGTCATTGACAT |
| gapC1-gRNA | CGATTTACAACTGGTGAAAA |
| gapC2-gRNA | CGTTGATGGGAAAAGTATCG |
| S9-gRNA | TCTGGCGCAGTTGATATGTA |
| P glk -gRNA | TCTCACACTGTAAATACCTG |
| P mak -gNRA | TAAATCGATACCTATACGCA |
| Cgeno-gRNA | AGGAAGTGAGTGAATTCTTCAGAGGTGAACGC |
| Lafu-gRNA | AAGCTCGGTGGCGGATTTCTTCTTCAGCAGCG |
| ydeU-gRNA | TGGTTGGCTGGTATGGCACTGGAGTGCTTAAT |
| ycdN-gRNA | GTTTCTCATTATGTTGCGCGAAGGACTTGAAG |
| adhE-gRNA | GGCATGGGTATCGTCGAAGATAAAGTGATCAA |
| ycjV-gRNA | GGGCTTGAGGAGATCAGCGGCGGCGATCTGTT |
| ycgh-gRNA | TTCAGATAAGGTGATGCAAGGATTGCAGCTGG |
| yciQ-gRNA | TGGTTTGTGGATGGTTATATCTCTGGAAGCGC |
Table 2 Primers and gRNA used in this work
引物名称 Primer name | 引物序列 Primer sequence (5′-3′) |
|---|---|
| gapC2-up-F | CACTAGTGAATTCGCGGCCGCCTGCAGAACAGGAGATAAATCACCAAATGTCCCAAATGCGC |
| gapC2-up-R | AGAGCTCTCCCATATGGTCGACATAAGTGAATCTTCCGTAAAATCAACGC |
| gapC2-down-F | TCGACCATATGGGAGAGCTCTTTACGCCGAAAAAGAGGCTAAAAATATTC |
| gapC2-down-R | AATACTCAAGCTATGCATCCAACGCGTTGGGTTTGCGTGGCATCAAACACCGAACCG |
| gRNA2-gapC-F | TTGACAGCTAGCTCAGTCCTAGGTATAATGCTAGCCGTTGATGGGAAAAGTATCGGTTTTAGAGCTAGAAATAGCAAGTT |
| gRNA2-gapC-R | GGTTCTTATGGCTCTTGTATCTATCAGTGAAGCATCAAGAC |
| S9-up-F | CACTAGTGAATTCGCGGCCGCCTGCAGGCACGGAGAATGGCAGGAAGCATC |
| S9-up-R | CAATCCTACCGAGCTCTCCCATATGGTCGACCAGGTTTATTATATCGCGTTGATTATTGATGCTGTTTTTAGTTTTAACGGC |
| S9-down-F | TAATAAACCTGGTCGACCATATGGGAGAGCTCGGTAGGATTGAAAACGCTCTCCTGATTTTCCAATTCA |
| S9-down-R | GCTATGCATCCAACGCGTTGGGCAGCCCGCCCGGTTGCCCG |
| gRNA-S9-F | TTGACAGCTAGCTCAGTCCTAGGTATAATGCTAGCTCTGGCGCAGTTGATATGTAGTTTTAGAGCTAGAAATAGCAAGTT |
| gRNA-S9-R | GGTTCTTATGGCTCTTGTATCTATCAGTGAAGCATCAAGAC |
| gapA-Scshb17-F | ACGGAAGATTCACTTATGTCGACATCTCGACGAAATGGCTGCAC |
| gapA-Scshb17-R | CATCTGGGGGTTAGCGAAGGCATATATTCCACCAGCTATTTGTTAGTG |
| Scshb17-F | CACTAACAAATAGCTGGTGGAATATATGCCTTCGCTAACCCCCAG |
| Scshb17-R | TTAGCCTCTTTTTCGGCGTAAAGAGCTCTTACACATCGCCATGCTGGG |
| GapA-fucP-F | CAATAATCAACGCGATATAATAAACCTGATCTCGACGAAATGGCTGCAC |
| GapA-fucP-R | GTTTGTATTGATGTGTTTCCCATATATTCCACCAGCTATTTGTTAG |
| fucP-F | ACAAATAGCTGGTGGAATATATGGGAAACACATCAATACAAACGC |
| fucP-R | CAGGAGAGCGTTTTCAATCCTACCGAGCTCCATGGATCCTCAGTTAGTTGCCGTTTGAGAACG |
| gRNA1-gapC-F | TTGACAGCTAGCTCAGTCCTAGGTATAATGCTAGCCGATTTACAACTGGTGAAAAGTTTTAGAGCTAGAAATAGCAAGTT |
| gRNA1-gapC-R | GGTTCTTATGGCTCTTGTATCTATCAGTGAAGCATCAAGAC |
| gapC1-up-F | CACTAGTGAATTCGCGGCCGCCTGCAGCTAAAAATATTCCGTGGAAAGCGAAAGGTGCAGAAAT |
| gapC1-up-R | CCATATTCGTTATCGTACCAGGCGAGAGCTCTCCCATATGGTCGACCCCACGGCGGTAATTTCCGTTTGC |
| gapC1-down-F | CGGAAATTACCGCCGTGGGGTCGACCATATGGGAGAGCTCTCGCCTGGTACGATAACGAATATGGCTTCG |
| gapC1-down-R | CTCAAGCTATGCATCCAACGCGTTGGTTTATGATCAGTTCGTCAATCGGCTGGGTGAAGC |
| phac1+glf-F | GAGCTCTAGCTGGACGTCTAGAGTTGACAGCGCGTGCGTTGCAAG |
| Glf-R | GCCATATTCGTTATCGTACCAGGCGATTATTTCTGGCTGCGCCACATTTC |
| gRNA-P glk -F | TTGACAGCTAGCTCAGTCCTAGGTATAATGCTAGCTCTCACACTGTAAATACCTGGTTTTAGAGCTAGAAATAGCAAGTT |
| gRNA-P glk -R | GGTTCTTATGGCTCTTGTATCTATCAGTGAAGCATCAAGAC |
| glk-up-F | ATTCGCGGCCGCCTGCAGGTCGACCTATTCGGCGCAAAATCAACGT |
| glk-up-R | GAAAGAGGAGAAATACTAGATGACAAAGTATGCATTAGTCGGTGATGTGGGC |
| glk-down-F | CTTTGTCATCATATGACGGAGCTCAGGCAGGCTCCCTGTAAATATC |
| glk-down-R | CAAGCTATGCATCCAACGCGTTGGCGGTCATGATCGGATGTTCCG |
| gRNA-P tktA -F | TTGACAGCTAGCTCAGTCCTAGGTATAATGCTAGCGATCGGATGATGAAGGGCACGTTTTAGAGCTAGAAATAGCAAGTT |
| gRNA-P tktA -R | GGTTCTTATGGCTCTTGTATCTATCAGTGAAGCATCAAGAC |
| gapA-eno-F | CATTTGAGAAGCACACGGTCACACTGCATCTCGACGAAATGGCTGCAC |
| gapA-eno-R | ACGTGCATGATTTCAGCCATATATTCCACCAGCTATTTGTTAGTGAATAAAAG |
| Cgeno-F | CACTAACAAATAGCTGGTGGAATATATGGCTGAAATCATGCACGTATTC |
| Cgeno-R | GTGCGTCGGGTGATGCTGCCAACTTACTTAGCCCTGAAAGCGTGGGAATG |
| CmR-F | GTATCCGCTCATGAATTAATTCCAATAAACCGGTAAACCAGCAATAG |
| CmR-R | GCATTCCCACGCTTTCAGGGCTAAGTAAGTTGGCAGCATCACCCG |
| Scips-F | GAAAGAGGAGAAATACTAGATGACAGAAGATAATATTGCTCCAATCAC |
| imp-R | GACCGTGTGCTTCTCAAATGTTAACGCTTCAGAGCGTCGC |
| P tktA -gRNA | GATCGGATGATGAAGGGCAC |
| pgi-gRNA | TGGCAGGCACTACAGAAACACTTCGATGAAAT |
| zwf-gRNA | GAGACGGCTGAATGCAGCAGTGTCATTGACAT |
| gapC1-gRNA | CGATTTACAACTGGTGAAAA |
| gapC2-gRNA | CGTTGATGGGAAAAGTATCG |
| S9-gRNA | TCTGGCGCAGTTGATATGTA |
| P glk -gRNA | TCTCACACTGTAAATACCTG |
| P mak -gNRA | TAAATCGATACCTATACGCA |
| Cgeno-gRNA | AGGAAGTGAGTGAATTCTTCAGAGGTGAACGC |
| Lafu-gRNA | AAGCTCGGTGGCGGATTTCTTCTTCAGCAGCG |
| ydeU-gRNA | TGGTTGGCTGGTATGGCACTGGAGTGCTTAAT |
| ycdN-gRNA | GTTTCTCATTATGTTGCGCGAAGGACTTGAAG |
| adhE-gRNA | GGCATGGGTATCGTCGAAGATAAAGTGATCAA |
| ycjV-gRNA | GGGCTTGAGGAGATCAGCGGCGGCGATCTGTT |
| ycgh-gRNA | TTCAGATAAGGTGATGCAAGGATTGCAGCTGG |
| yciQ-gRNA | TGGTTTGTGGATGGTTATATCTCTGGAAGCGC |

Fig. 2 Phenotype verification of recombinant strain JY1 and JY2A: Growth curves of JY1 strain cultured in the medium containing different carbon sources. 10G: Fermentation medium contains 10.0 g/L glucose. 10F: Fermentation medium contains 10.0 g/L fructose. 5G5F: Fermentation medium contains 5.0 g/L glucose and 5.0 g/L fructose. B: Growth curve and inositol production of JY2 strain cultured in the medium containing mixed carbon source
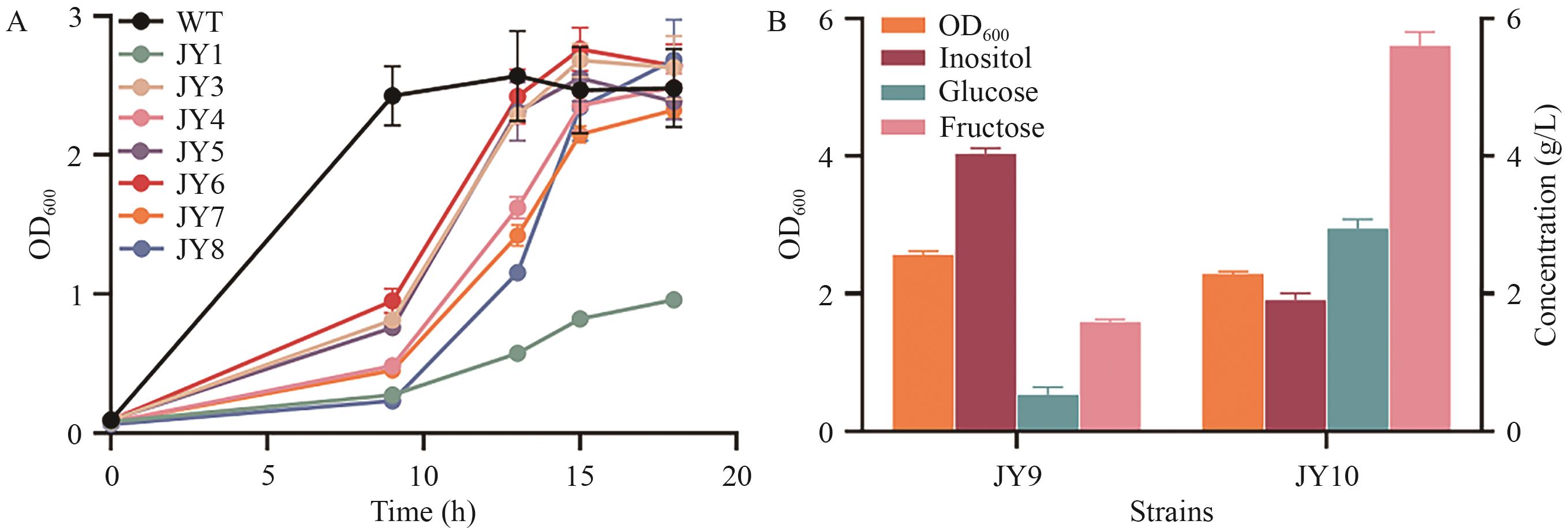
Fig. 3 Phenotype verification of recombinant strain JY3-JY10A: Growth curves of recombinant strains cultured in the medium containing mixed carbon source. B: Inositol production, carbon source concentration and maximum OD600 of recombinant strains cultured in the medium containing mixed carbon source

Fig. 4 Fermentation results of recombinant strains JY12-JY14 in 3 L reactorA: Fermentation results of JY12 strain in 3 L reactor. B: Fermentation results of JY13 strain in 3 L reactor. C: Fermentation results of JY14 strain in 3 L reactor
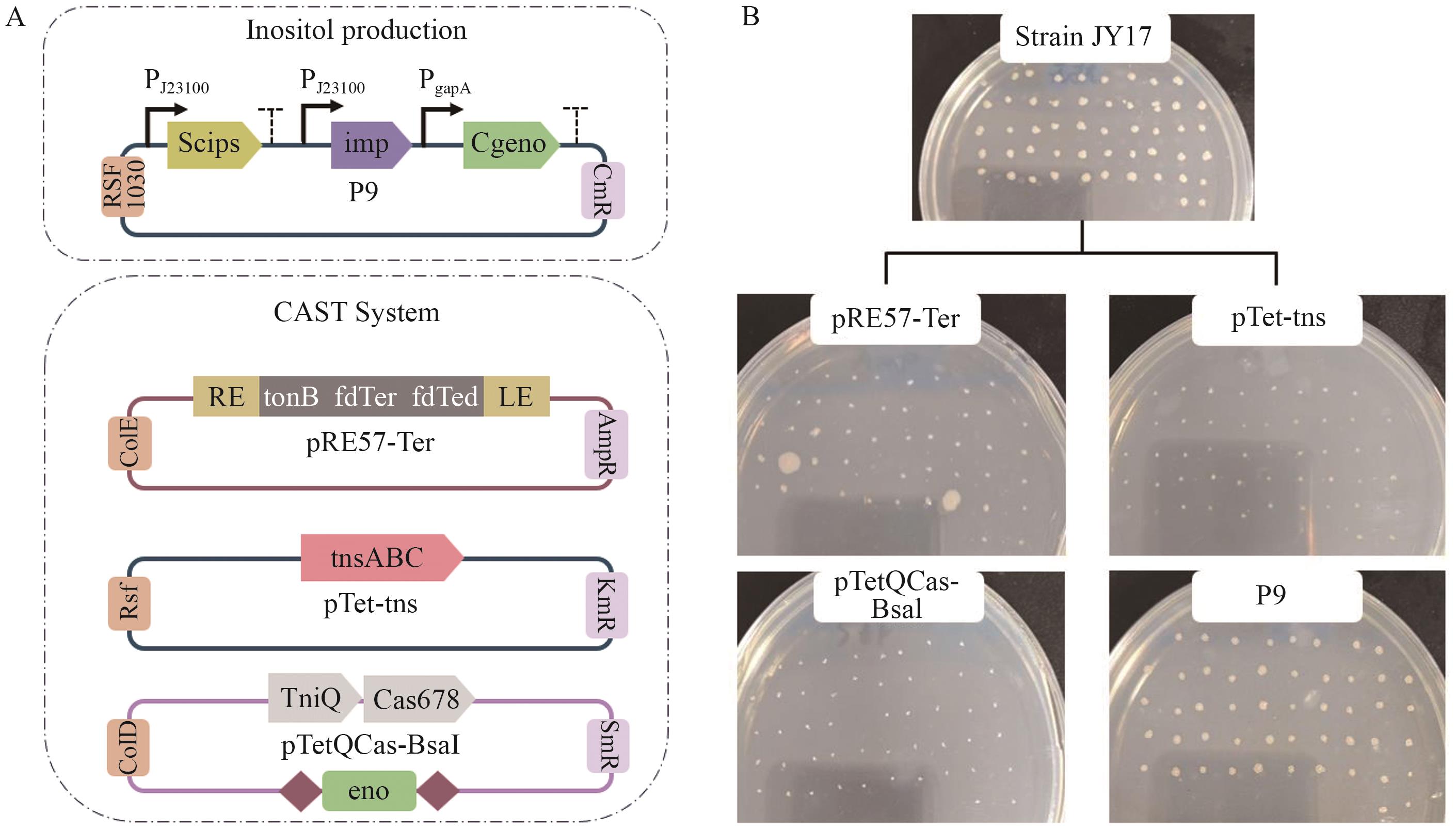
Fig. 5 Establishment and stability evaluation of Scips, imp and eno synergetic expression systemA: P9 plasmid map and CAST system. B: Stability verification of gene editing and P9 expression plasmids
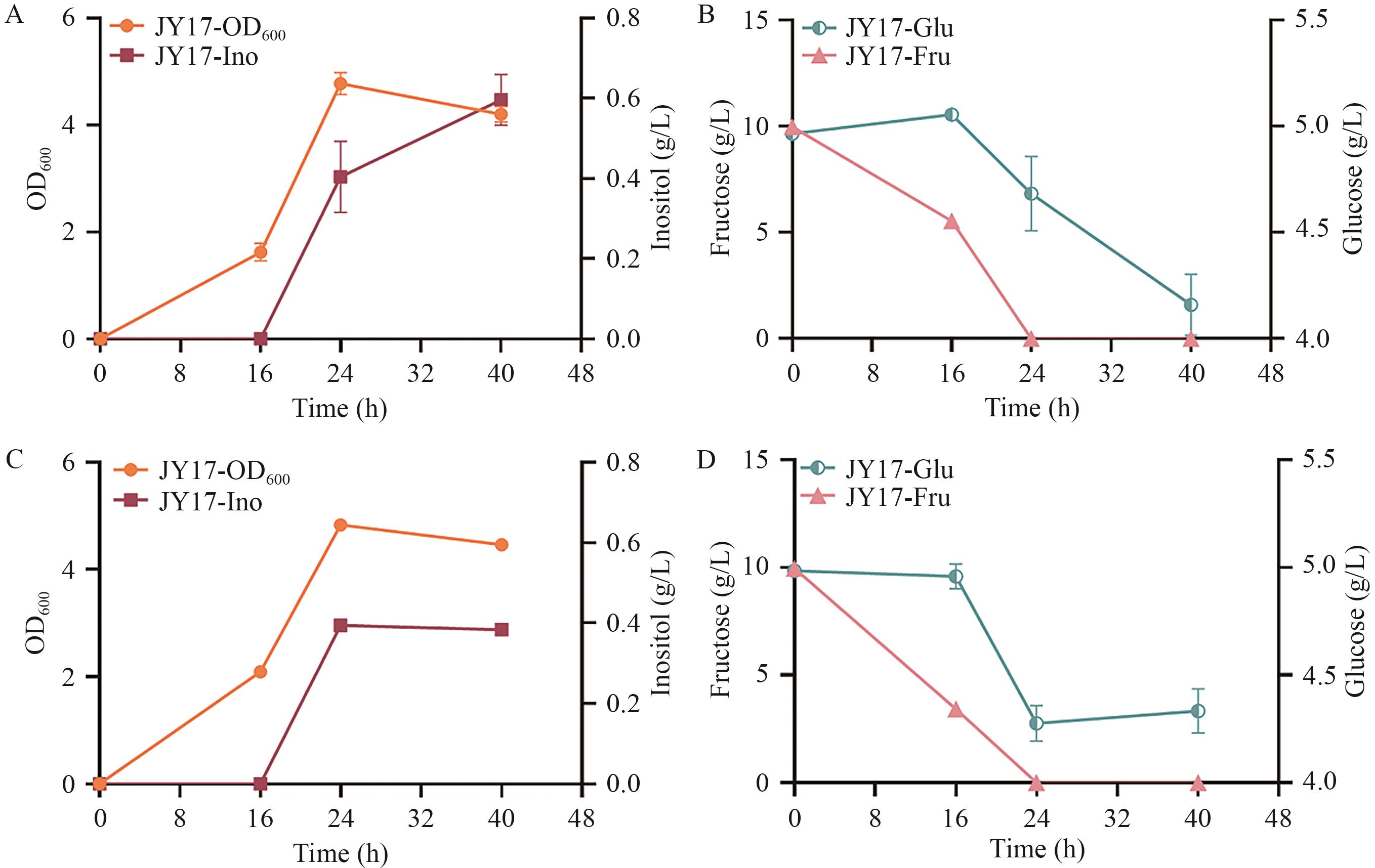
Fig. 6 Shake flask fermentation results of recombinant strain JY17A: Growth curve and inositol production during fermentation without adding antibiotics. B: Glucose and fructose consumption during fermentation without adding antibiotic. C: Growth curve and inositol production during fermentation with adding antibiotic. D: Glucose and fructose consumption during fermentation with adding antibiotic
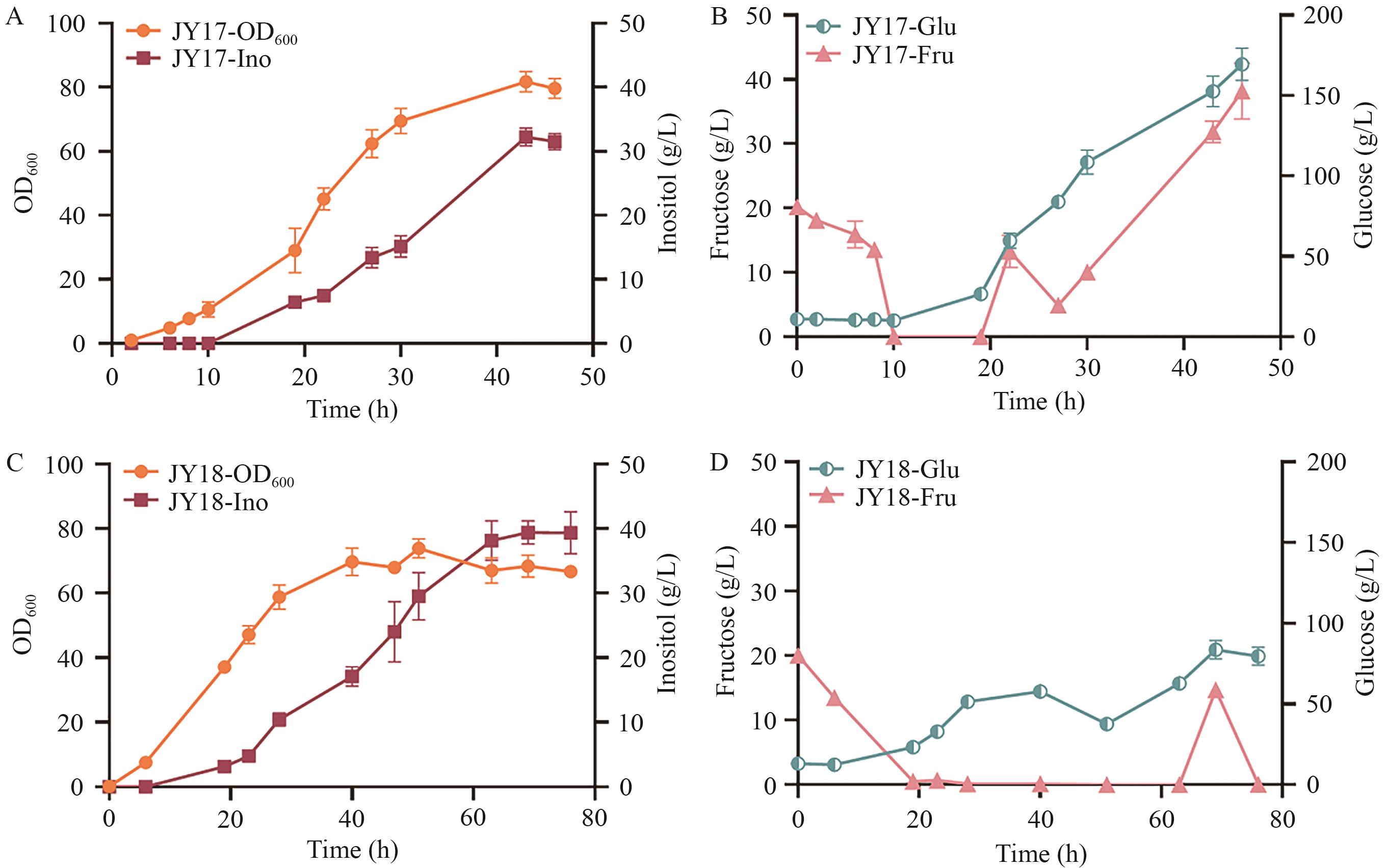
Fig. 7 Fermentated results of recombinant strain JY17 and JY18 in 3 L reactorA: Growth curve and inositol production of JY17. B: Glucose and fructose consumption of JY17. C: Growth curve and inositol production of JY18. D: Glucose and fructose consumption of JY18
| [1] | Murthy PPN. Structure and nomenclature of inositol phosphates, phosphoinositides, and glycosylphosphatidylinositols [J]. Subcell Biochem, 2006, 39: 1-19. |
| [2] | Sinclair KD, Allegrucci C, Singh R, et al. DNA methylation, insulin resistance, and blood pressure in offspring determined by maternal periconceptional B vitamin and methionine status [J]. Proc Natl Acad Sci USA, 2007, 104(49): 19351-19356. |
| [3] | 张泽生, 董硕, 高云峰, 等. 肌醇、D-手性肌醇对大鼠酒精性脂肪肝作用的研究 [J]. 中国食品添加剂, 2014, 25(9): 85-90. |
| Zhang ZS, Dong S, Gao YF, et al. Effect of inositol and D-chiro-inositol on alcoholic fatty liver rats [J]. China Food Addit, 2014, 25(9): 85-90. | |
| [4] | Druzijanic A, Kovic M, Roguljic M, et al. Application of inositol hexaphosphate and inositol in dental medicine: an overview [J]. Biomolecules, 2023, 13(6): 913. |
| [5] | Özturan A, Arslan S, Kocaadam B, et al. Effect of inositol and its derivatives on diabetes: a systematic review [J]. Crit Rev Food Sci Nutr, 2019, 59(7): 1124-1136. |
| [6] | Etrusco A, Laganà AS, Chiantera V, et al. Myo-inositol in assisted reproductive technology from bench to bedside [J]. Trends Endocrinol Metab, 2024, 35(1): 74-83. |
| [7] | Sharma N, Watkins OC, Chu AHY, et al. Myo-inositol: a potential prophylaxis against premature onset of labour and preterm birth [J]. Nutr Res Rev, 2023, 36(1): 60-68. |
| [8] | Heimark D, McAllister J, Larner J. Decreased myo-inositol to chiro-inositol (M/C) ratios and increased M/C epimerase activity in PCOS theca cells demonstrate increased insulin sensitivity compared to controls [J]. Endocr J, 2014, 61(2): 111-117. |
| [9] | López-Gambero AJ, Sanjuan C, Serrano-Castro PJ, et al. The biomedical uses of inositols: a nutraceutical approach to metabolic dysfunction in aging and neurodegenerative diseases [J]. Biomedicines, 2020, 8(9): 295. |
| [10] | Michell RH. Do inositol supplements enhance phosphatidylinositol supply and thus support endoplasmic reticulum function? [J]. Br J Nutr, 2018, 120(3): 301-316. |
| [11] | Gupta A, Sanwal N, Bareen MA, et al. Trends in functional beverages: Functional ingredients, processing technologies, stability, health benefits, and consumer perspective [J]. Food Res Int, 2023, 170: 113046. |
| [12] | Yan EF, Sun HJ, He LJ, et al. Dietary inositol supplementation improves meat quality by modulating amino acid metabolism and gut microbiota composition of finishing pigs [J]. Anim Nutr, 2024, 19: 180-191. |
| [13] | Shiau SY, Su SL. Dietary inositol requirement for juvenile grass shrimp, Penaeus monodon [J]. Aquaculture, 2004, 241(1-4): 1-8. |
| [14] | Rodrigues EJD, de Carvalho PLPF, Silva VF, et al. Myo-inositol increases respiratory burst, cellular proliferation, and phagocytosis of cultured leukocytes from Nile Tilapia [J]. Comp Immunol Rep, 2025, 8: 200193. |
| [15] | Li B, Pan SM, Huang WB, et al. The positive effects of dietary inositol on juvenile hybrid grouper (♀Epinephelus fuscoguttatus × ♂ Epinephelus lanceolatu) fed high-lipid diets: Growth performance, intestinal digestive enzymes, tissue morphology, and intestinal microbiota [J]. Aquac Rep, 2024, 39: 102534. |
| [16] | Michael FR, Koshio S. Biochemical studies on the interactive effects of dietary choline and inositol in juvenile Kuruma shrimp, Marsupenaeus japonicus Bate [J]. Aquaculture, 2008, 285(1-4): 179-183. |
| [17] | Febles CI, Arias A, Hardisson A, et al. Phytic acid level in wheat flours [J]. J Cereal Sci, 2002, 36(1): 19-23. |
| [18] | 黄贞杰, 陈由强, 陈丽霞, 等. 代谢工程改造酿酒酵母合成肌醇 [J]. 微生物学通报, 2017, 44(10): 2289-2296. |
| Huang ZJ, Chen YQ, Chen LX, et al. Metabolic engineering of Saccharomyces cerevisiae for inositol production [J]. Microbiol China, 2017, 44(10): 2289-2296. | |
| [19] | Zhang QQ, Wang XL, Luo HY, et al. Metabolic engineering of Pichia pastoris for myo-inositol production by dynamic regulation of central metabolism [J]. Microb Cell Fact, 2022, 21(1): 112. |
| [20] | You R, Wang L, Shi CR, et al. Efficient production of myo-inositol in Escherichia coli through metabolic engineering [J]. Microb Cell Fact, 2020, 19(1): 109. |
| [21] | 邹世能. 甘油生产的状况和未来 [J]. 日用化学工业, 1996, 26(3): 36-38. |
| Zou SN. Present situation and future of glycerol production [J]. China Surfactant Deterg Cosmet, 1996, 26(3): 36-38. | |
| [22] | Khanna S, Goyal A, Moholkar VS. Microbial conversion of glycerol: present status and future prospects [J]. Crit Rev Biotechnol, 2012, 32(3): 235-262. |
| [23] | Kuznetsova E, Xu LD, Singer A, et al. Structure and activity of the metal-independent fructose-1, 6-bisphosphatase YK23 from Saccharomyces cerevisiae [J]. J Biol Chem, 2010, 285(27): 21049-21059. |
| [24] | Hellgren J, Godina A, Nielsen J, et al. Promiscuous phosphoketolase and metabolic rewiring enables novel non-oxidative glycolysis in yeast for high-yield production of acetyl-CoA derived products [J]. Metab Eng, 2020, 62: 150-160. |
| [25] | Zhang YW, Yang JW, Yang SQ, et al. Programming cells by multicopy chromosomal integration using CRISPR-associated transposases [J]. CRISPR J, 2021, 4(3): 350-359. |
| [26] | Li Q, Sun BB, Chen J, et al. A modified pCas/pTargetF system for CRISPR-Cas9-assisted genome editing in Escherichia coli [J]. Acta Biochim Biophys Sin, 2021, 53(5): 620-627. |
| [27] | Shiue E, Brockman IM, Prather KLJ. Improving product yields on D-glucose in Escherichia coli via knockout of pgi and zwf and feeding of supplemental carbon sources [J]. Biotechnol Bioeng, 2015, 112(3): 579-587. |
| [28] | Li YJ, Han PP, Wang J, et al. Production of myo-inositol: recent advance and prospective [J]. Biotechnol Appl Biochem, 2022, 69(3): 1101-1111. |
| [29] | Li BB, Wang X, Tai L, et al. NAD kinases: metabolic targets controlling redox co-enzymes and reducing power partitioning in plant stress and development [J]. Front Plant Sci, 2018, 9: 379. |
| [30] | Kornberg H, Lourenco C. A route for fructose utilization by Escherichia coli involving the fucose regulon [J]. Proc Natl Acad Sci USA, 2006, 103(51): 19496-19499. |
| [31] | Ji H, Wang JF, Guo JR, et al. Progress in the biological function of alpha-enolase [J]. Anim Nutr, 2016, 2(1): 12-17. |
| [32] | Usui Y, Hirasawa T, Furusawa C, et al. Investigating the effects of perturbations to pgi and eno gene expression on central carbon metabolism in Escherichia coli using 13C metabolic flux analysis [J]. Microb Cell Fact, 2012, 11: 87. |
| [1] | YAN Meng-yang, LIANG Xiao-yang, DAI Jun-ang, ZHANG Yan, GUAN Tuan, ZHANG Hui, LIU Liang-bo, SUN Zhi-hua. Screening of Amoxicillin-degrading Bacteria and Study on Its Degradation Mechanisms [J]. Biotechnology Bulletin, 2025, 41(9): 314-325. |
| [2] | HUANG Xu-sheng, ZHOU Ya-li, CHAI Xu-dong, WEN Jing, WANG Ji-ping, JIA Xiao-yun, LI Run-zhi. Cloning of Plastidial PfLPAT1B Gene from Perilla frutescens and Its Functional Analysis in Oil Biosynthesis [J]. Biotechnology Bulletin, 2025, 41(7): 226-236. |
| [3] | LU Tian-yi, LI Ai-peng, FEI Qiang. Research Progress in the Biosynthesis of Polylactic Acid [J]. Biotechnology Bulletin, 2025, 41(4): 47-60. |
| [4] | WEI Min-hua, LI Xiao-tong, JIANG Ya-wen, ZHOU Piao-piao, WANG Kai, SUN Hao, LU Nan, ZHANG Cheng-lin. Systems Metabolic Engineering for Highly Efficient L-isoleucine Production in Escherichia coli [J]. Biotechnology Bulletin, 2025, 41(11): 110-120. |
| [5] | RAO Jun, ZHAO Chen, LI Duan-hua, LIAO Hao, HUANG Jia-yu, WANG Lu. Application of Auto-induction Strategy in Ergothioneine Biosynthesis [J]. Biotechnology Bulletin, 2025, 41(1): 333-346. |
| [6] | ZHANG Jing-an, HU Xiao-long, CAO Bei-bei, LIAO Min, SHU Chang-long, ZHANG Jie, WANG Kui, CAO Hai-qun. Construction and Characterization of Rapid Visual Expression Vector for Bacillus thuringiensis [J]. Biotechnology Bulletin, 2025, 41(1): 95-102. |
| [7] | ZHANG Mei-yu, ZHAO Yu-bin, WANG Ling-yun, SONG Yuan-da, ZHAO Xin-he, REN Xiao-jie. Research Progress in the Production of Functional Fatty Acid DHA by Microalga Thraustochytrids [J]. Biotechnology Bulletin, 2024, 40(6): 81-94. |
| [8] | WANG Zhou, YU Jie, WANG Jin-hua, WANG Yong-ze, ZHAO Xiao. Anaerobic Expression of Lactate Dehydrogenase to Improve the D-lactic Acid Optical Purity Procluced by Escherichia coli [J]. Biotechnology Bulletin, 2024, 40(5): 290-299. |
| [9] | ZHUANG Ke, LIANG Zhi-xuan, HE Ying-ting, XIE Qiu-ling. Transfer of Antibiotic-resistance Gene AmpR by Escherichia coli DH5α Through Outer Membrane Vesicles [J]. Biotechnology Bulletin, 2024, 40(12): 275-281. |
| [10] | HE Si-cheng, ZHANG Zi-yuan, HAN Yu-qing, MIAO Lin, ZHANG Cui-ying, YU Ai-qun. Research Progress in the Production of Polyunsaturated Fatty Acids by Yarrowia lipolytica Cell Factories [J]. Biotechnology Bulletin, 2024, 40(1): 72-85. |
| [11] | LI Liang, XU Shan-shan, JIANG Yan-jun. Research Progress in the Production of Ergothioneine by Biosynthesis [J]. Biotechnology Bulletin, 2024, 40(1): 86-99. |
| [12] | YANG Hong-yan, HAN Xiao, YANG Jian-jun. Scaling up Production of pDNA Plasmids in Disposable Bioreactors [J]. Biotechnology Bulletin, 2024, 40(1): 168-175. |
| [13] | XUE Ning, WANG Jin, LI Shi-xin, LIU Ye, CHENG Hai-jiao, ZHANG Yue, MAO Yu-feng, WANG Meng. Construction of L-phenylalanine High-producing Corynebacterium glutamicum Engineered Strains via Multi-gene Simultaneous Regulation Combined with High-throughput Screening [J]. Biotechnology Bulletin, 2023, 39(9): 268-280. |
| [14] | CHENG Ya-nan, ZHANG Wen-cong, ZHOU Yuan, SUN Xue, LI Yu, LI Qing-gang. Synthetic Pathway Construction of Producing 2'-fucosyllactose by Lactococcus lactis and Optimization of Fermentation Medium [J]. Biotechnology Bulletin, 2023, 39(9): 84-96. |
| [15] | ZHAO Si-jia, WANG Xiao-lu, SUN Ji-lu, TIAN Jian, ZHANG Jie. Modification of Pichia pastoris for Erythritol Production by Metabolic Engineering [J]. Biotechnology Bulletin, 2023, 39(8): 137-147. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||