








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (11): 110-120.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0695
WEI Min-hua( ), LI Xiao-tong, JIANG Ya-wen, ZHOU Piao-piao, WANG Kai, SUN Hao, LU Nan, ZHANG Cheng-lin(
), LI Xiao-tong, JIANG Ya-wen, ZHOU Piao-piao, WANG Kai, SUN Hao, LU Nan, ZHANG Cheng-lin( )
)
Received:2025-06-30
Online:2025-11-26
Published:2025-12-09
Contact:
ZHANG Cheng-lin
E-mail:22914013@mail.tust.edu.cn;zcl@tust.edu.cn
WEI Min-hua, LI Xiao-tong, JIANG Ya-wen, ZHOU Piao-piao, WANG Kai, SUN Hao, LU Nan, ZHANG Cheng-lin. Systems Metabolic Engineering for Highly Efficient L-isoleucine Production in Escherichia coli[J]. Biotechnology Bulletin, 2025, 41(11): 110-120.
底盘细胞 Chassis | 主要策略 Main strategies | 产量 Titer (g/L) | 转化率 Yield (g/g Glucose) | 周期 Period(h) | 发酵强度Productivity(g/L/h) | 参考文献 Reference |
|---|---|---|---|---|---|---|
谷氨酸棒杆菌 Corynebacterium glutamicum | 敲除alaT、brnQ和alr;过表达ilvBNM、ppnK、lrp、brnFE和ilvAM | 32.1 | 0.18 | 72 | 0.45 | [ |
大肠杆菌 Escherichia coli | 过表达ilvAfbr、ilvIH、thrABC、ygaZH、ilvEDA和ilvC | 9.5 | 0.14 | 60 | 0.16 | [ |
| 过表达ilvIH1、ygaZH和CgilvA1;敲除poxB、pflB、ldhA、adhE和tdcC | 49.3 | 0.32 | 48 | 1.03 | [ | |
| 敲除brnQ、livj、livk和ackA;过表达ygaZH、cimA3.7、GsleuCD、AfleuB ilvIHfbr、ilvCNADH、ilvD、LsleuDH和dcuD; | 56.6 | 0.21 | 34 | 1.66 | [ | |
| 过表达ilvAYI、ilvBNYI、thrAfbrBC、pntAB、aspA、brnFE、bcd、metAG189C和metBM;敲除brnQ | 51.5 | 0.29 | 44 | 1.13 | [ | |
| 过表达thrAfbrBC、ilvGM(AT)、asd、aspA、ilvEDAfbr、rhtC和pntAB;敲除pykF和ptsG | 5.42 | 0.40 | 12 | 0.83 | [ |
Table 1 Recent advances in metabolic engineering for L-isoleucine production
底盘细胞 Chassis | 主要策略 Main strategies | 产量 Titer (g/L) | 转化率 Yield (g/g Glucose) | 周期 Period(h) | 发酵强度Productivity(g/L/h) | 参考文献 Reference |
|---|---|---|---|---|---|---|
谷氨酸棒杆菌 Corynebacterium glutamicum | 敲除alaT、brnQ和alr;过表达ilvBNM、ppnK、lrp、brnFE和ilvAM | 32.1 | 0.18 | 72 | 0.45 | [ |
大肠杆菌 Escherichia coli | 过表达ilvAfbr、ilvIH、thrABC、ygaZH、ilvEDA和ilvC | 9.5 | 0.14 | 60 | 0.16 | [ |
| 过表达ilvIH1、ygaZH和CgilvA1;敲除poxB、pflB、ldhA、adhE和tdcC | 49.3 | 0.32 | 48 | 1.03 | [ | |
| 敲除brnQ、livj、livk和ackA;过表达ygaZH、cimA3.7、GsleuCD、AfleuB ilvIHfbr、ilvCNADH、ilvD、LsleuDH和dcuD; | 56.6 | 0.21 | 34 | 1.66 | [ | |
| 过表达ilvAYI、ilvBNYI、thrAfbrBC、pntAB、aspA、brnFE、bcd、metAG189C和metBM;敲除brnQ | 51.5 | 0.29 | 44 | 1.13 | [ | |
| 过表达thrAfbrBC、ilvGM(AT)、asd、aspA、ilvEDAfbr、rhtC和pntAB;敲除pykF和ptsG | 5.42 | 0.40 | 12 | 0.83 | [ |
菌株/质粒 Strains/Plasmids | 特性 Characteristics | 来源 Sources |
|---|---|---|
| 菌株 | ||
| E. coli DH5α | F-, Δ(lacZYA-argF)U169 recA1endA1 hsdR17 | 实验室保存 |
| ISO-2 | E. coli W3110 ΔlacI P thrABC -thrA::P trc -thrAfbrylbE::P trc -thrAfbrBC-T trcyjiP::P trc -ilvAYI-T trcyncI::P trc -ilvBNYI-T trc | [ |
| YL-1 | ISO-2 tehB::P trc -ppc | 本研究 |
| YL-2 | YL-1 yciQ::P trc -pycA | 本研究 |
| YL-3 | YL-2 yeeP::P trc -aspC | 本研究 |
| YL-4 | YL-3 ygaY::P trc -aspA | 本研究 |
| YL-5 | YL-4 yjgX::P trc -ilvAYI | 本研究 |
| YL-6 | YL-5 yghX::P trc -ilvD | 本研究 |
| YL-7 | YL-6 P ilvC -ilvC::P trc -ilvCEM | 本研究 |
| YL-8 | YL-7 P ilvE -ilvE::P trc -bcd | 本研究 |
| YL-9 | YL-8 yeeL::P tr -leuBCD | 本研究 |
| YL-10 | YL-9 gapC::PBBa_J23110-cimA | 本研究 |
| YL-11 | YL-9 gapC::PBBa_J23104-cimA | 本研究 |
| YL-12 | YL-9 gapC::PBBa_J23119-cimA | 本研究 |
| YL-13 | YL-12 △iclR | 本研究 |
| YL-14 | YL-13 P sucAB ::P fliA | 本研究 |
| YL-15 | YL-14 △brnQ | 本研究 |
| YL-16 | YL-15 yjiV::P trc -ygaZH | 本研究 |
| 质粒 | ||
| pREDCas9 | SpeR,Cas9和λ Red表达质粒 | [ |
| pGRB | AmpR,gRNA表达质粒 | [ |
| pGRB-tehB | pGRB含tehB靶点sgRNA | 本研究 |
| pGRB-yciQ | pGRB含yciQ靶点sgRNA | 本研究 |
| pGRB-yeeP | pGRB含yeeP靶点sgRNA | 本研究 |
| pGRB-ygaY | pGRB含ygaY靶点sgRNA | 本研究 |
| pGRB-yjgX | pGRB含yjgX靶点sgRNA | 本研究 |
| pGRB-yghX | pGRB含yghX靶点sgRNA | 本研究 |
| pGRB-ilvC | pGRB含ilvC靶点sgRNA | 本研究 |
| pGRB-ilvE | pGRB含ilvE靶点sgRNA | 本研究 |
| pGRB-yeeL | pGRB含yeeL靶点sgRNA | 本研究 |
| pGRB-gapC | pGRB含gapC靶点sgRNA | 本研究 |
| pGRB-iclR | pGRB含iclR靶点sgRNA | 本研究 |
| pGRB-P sucAB | pGRB含P sucAB 靶点sgRNA | 本研究 |
| pGRB-brnQ | pGRB含brnQ靶点sgRNA | 本研究 |
| pGRB-yjiV | pGRB含yjiV靶点sgRNA | 本研究 |
Table 2 Strains and plasmids
菌株/质粒 Strains/Plasmids | 特性 Characteristics | 来源 Sources |
|---|---|---|
| 菌株 | ||
| E. coli DH5α | F-, Δ(lacZYA-argF)U169 recA1endA1 hsdR17 | 实验室保存 |
| ISO-2 | E. coli W3110 ΔlacI P thrABC -thrA::P trc -thrAfbrylbE::P trc -thrAfbrBC-T trcyjiP::P trc -ilvAYI-T trcyncI::P trc -ilvBNYI-T trc | [ |
| YL-1 | ISO-2 tehB::P trc -ppc | 本研究 |
| YL-2 | YL-1 yciQ::P trc -pycA | 本研究 |
| YL-3 | YL-2 yeeP::P trc -aspC | 本研究 |
| YL-4 | YL-3 ygaY::P trc -aspA | 本研究 |
| YL-5 | YL-4 yjgX::P trc -ilvAYI | 本研究 |
| YL-6 | YL-5 yghX::P trc -ilvD | 本研究 |
| YL-7 | YL-6 P ilvC -ilvC::P trc -ilvCEM | 本研究 |
| YL-8 | YL-7 P ilvE -ilvE::P trc -bcd | 本研究 |
| YL-9 | YL-8 yeeL::P tr -leuBCD | 本研究 |
| YL-10 | YL-9 gapC::PBBa_J23110-cimA | 本研究 |
| YL-11 | YL-9 gapC::PBBa_J23104-cimA | 本研究 |
| YL-12 | YL-9 gapC::PBBa_J23119-cimA | 本研究 |
| YL-13 | YL-12 △iclR | 本研究 |
| YL-14 | YL-13 P sucAB ::P fliA | 本研究 |
| YL-15 | YL-14 △brnQ | 本研究 |
| YL-16 | YL-15 yjiV::P trc -ygaZH | 本研究 |
| 质粒 | ||
| pREDCas9 | SpeR,Cas9和λ Red表达质粒 | [ |
| pGRB | AmpR,gRNA表达质粒 | [ |
| pGRB-tehB | pGRB含tehB靶点sgRNA | 本研究 |
| pGRB-yciQ | pGRB含yciQ靶点sgRNA | 本研究 |
| pGRB-yeeP | pGRB含yeeP靶点sgRNA | 本研究 |
| pGRB-ygaY | pGRB含ygaY靶点sgRNA | 本研究 |
| pGRB-yjgX | pGRB含yjgX靶点sgRNA | 本研究 |
| pGRB-yghX | pGRB含yghX靶点sgRNA | 本研究 |
| pGRB-ilvC | pGRB含ilvC靶点sgRNA | 本研究 |
| pGRB-ilvE | pGRB含ilvE靶点sgRNA | 本研究 |
| pGRB-yeeL | pGRB含yeeL靶点sgRNA | 本研究 |
| pGRB-gapC | pGRB含gapC靶点sgRNA | 本研究 |
| pGRB-iclR | pGRB含iclR靶点sgRNA | 本研究 |
| pGRB-P sucAB | pGRB含P sucAB 靶点sgRNA | 本研究 |
| pGRB-brnQ | pGRB含brnQ靶点sgRNA | 本研究 |
| pGRB-yjiV | pGRB含yjiV靶点sgRNA | 本研究 |
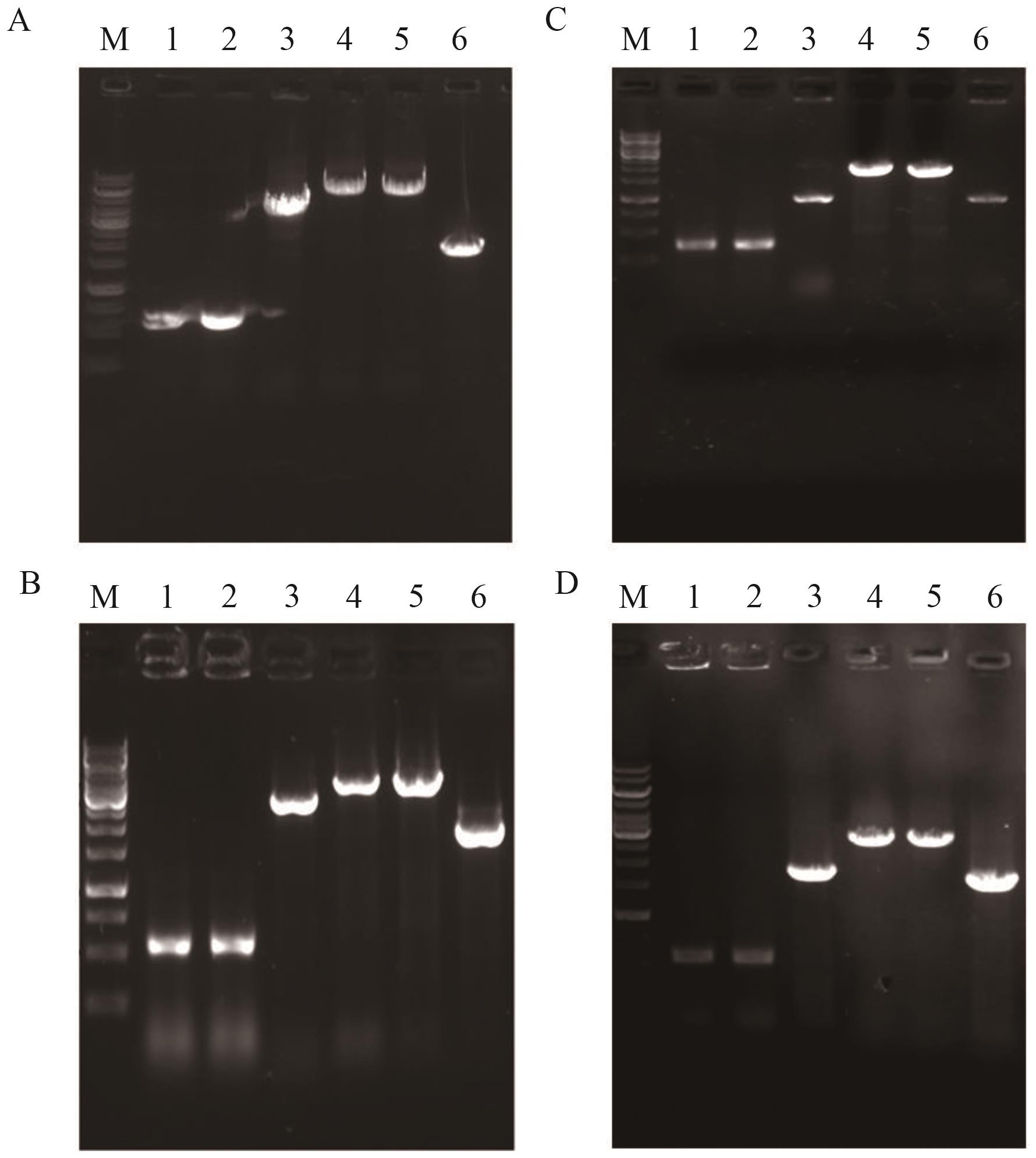
Fig. 2 Identification maps of YL-1, YL-2, YL-3 and YL-4A-D: Identification maps of YL-1, YL-2, YL-3 and YL-4. M: DNA marker (10 000, 8 000, 7 000, 6 000, 5 000, 40 00, 3 500, 3 000, 25 00, 2 000, 1 500, 1 000, 750, 500, 250 bp); 1: upstream homology arm; 2: downstream homology arm; 3: target gene; 4: overlapped PCR product; 5: PCR product amplified from genomic DNA of YL-1, YL-2, YL-3 and YL-4, respectively; 6: PCR product amplified from genomic DNA of the starting strain
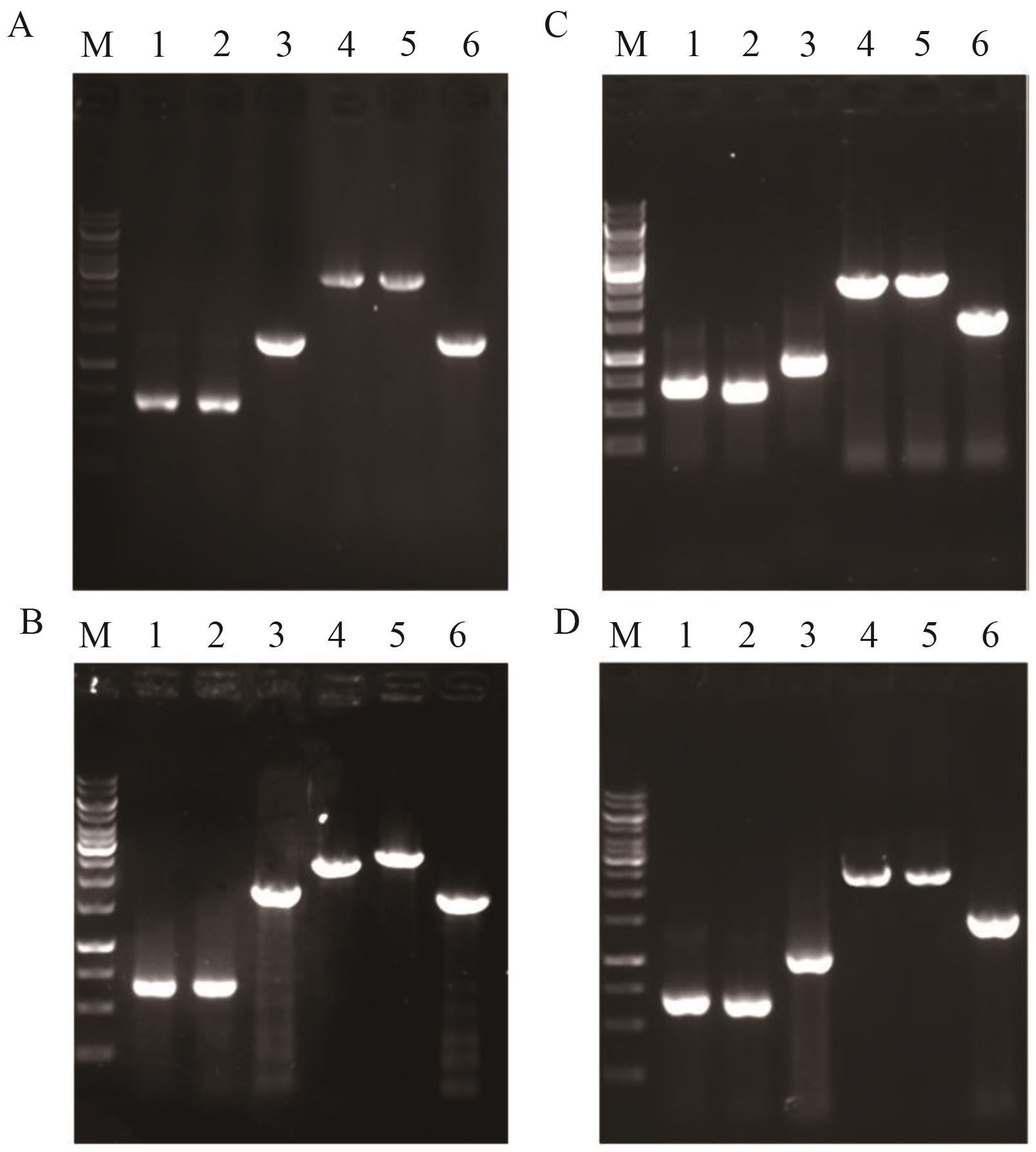
Fig. 4 Identification maps of YL-5-YL-8A-D: Identification maps of YL-5, YL-6, YL-7 and YL-8. M: DNA marker; 1: upstream homology arm; 2: downstream homology arm; 3: target gene; 4: overlap PCR product; 5: PCR product amplified from genomic DNA of YL-5, YL-6, YL-7 and YL-8, respectively; 6: PCR product amplified from genomic DNA of the starting strain
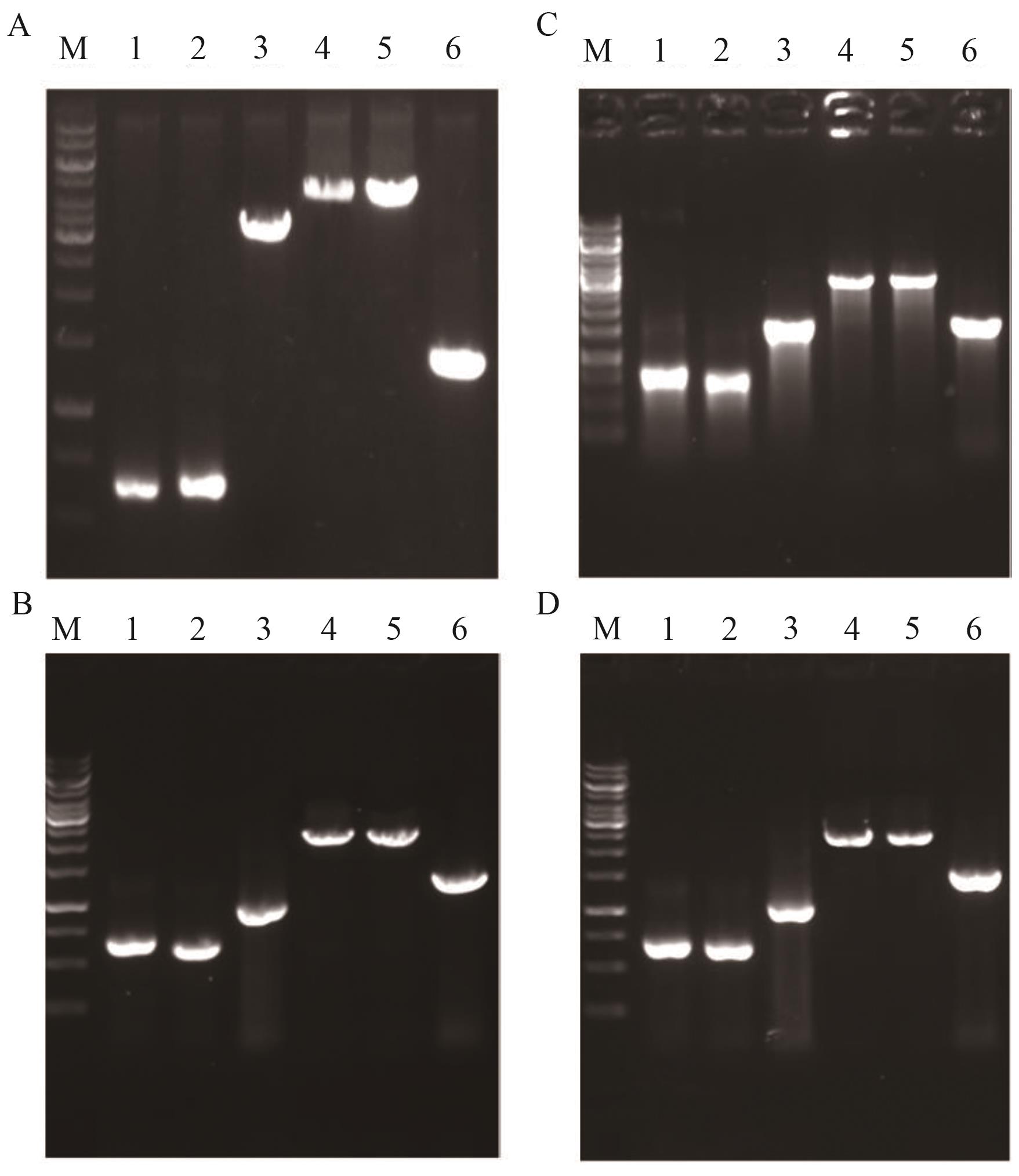
Fig. 6 Identification maps of YL-9-YL-12A-D: Identification maps of YL-9, YL-10, YL-11 and YL-12; M: DNA marker; 1: upstream homology arm; 2: downstream homology arm; 3: target gene; 4: overlap PCR product; 5: PCR product amplified from genomic DNA of YL-9, YL-10, YL-11 and YL-12, respectively; 6: PCR product amplified from genomic DNA of the starting strain

Fig. 7 Effects of introducing the citramalate pathway on strains' performancesA: L-Isoleucine production and biomass of strain YL-9, YL-10, YL-11, and YL-12. B: Intracellular α-ketobutyrate concentration in strain YL-8, YL-9, YL-10, YL-11, and YL-12
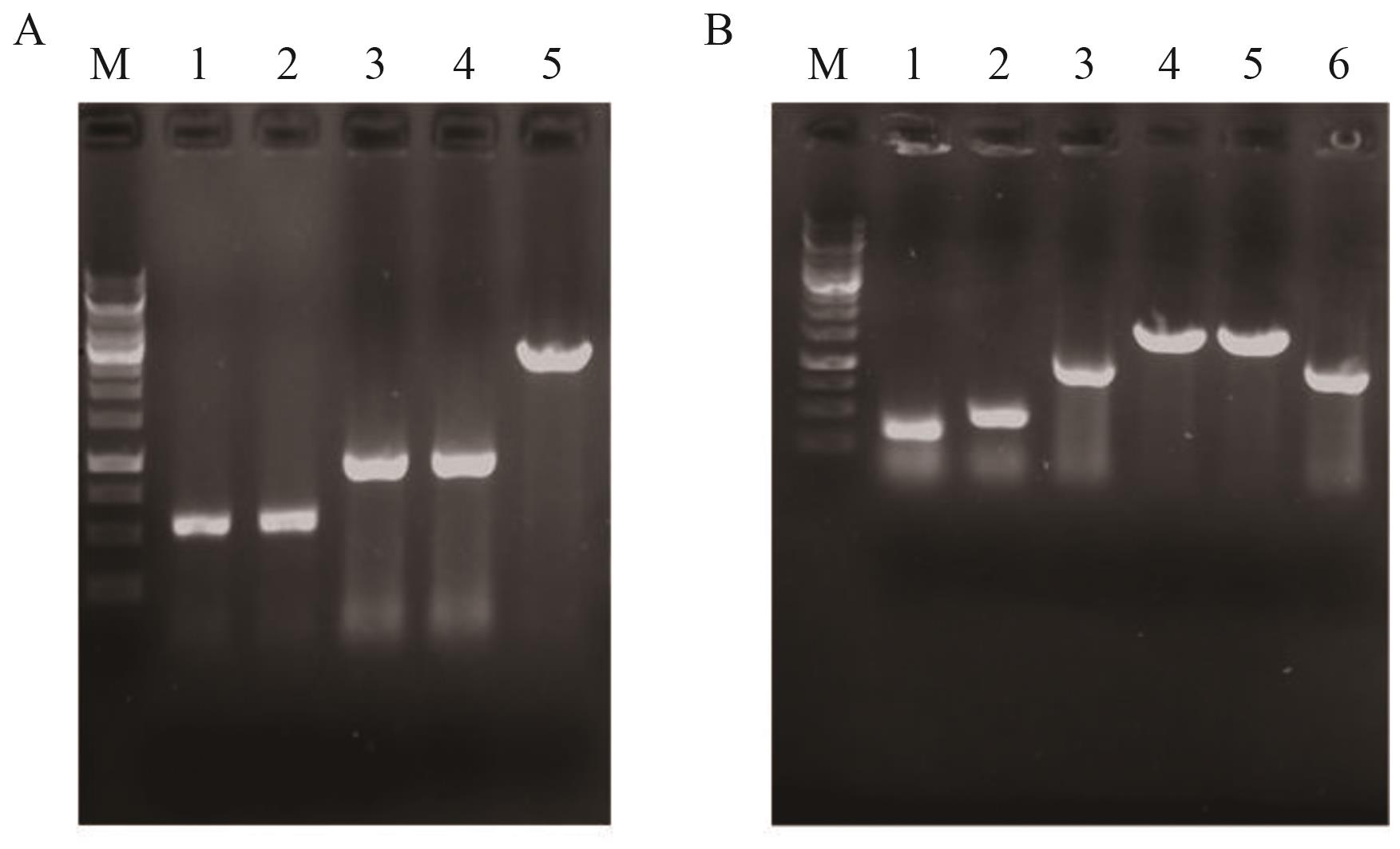
Fig. 8 Identification maps of YL-13 and YL-14A: Identification map of YL-13; M: DNA marker; 1: upstream homology arm U iclR; 2: downstream homology arm D iclR; 3: U iclR -D iclR; 4: PCR product amplified from YL-13 genomic DNA; 5: PCR product from YL-12 genomic DNA. B: Identification map of YL-14; M: DNA marker; 1: upstream homology arm UPsucAB; 2: downstream homology arm DPsucAB; 3: P fliA; 4: UPsucAB -P fliA -DPsucAB overlap PCR product; 5: PCR product amplified from YL-14 genomic DNA; 6: PCR product from YL-13 genomic DNA
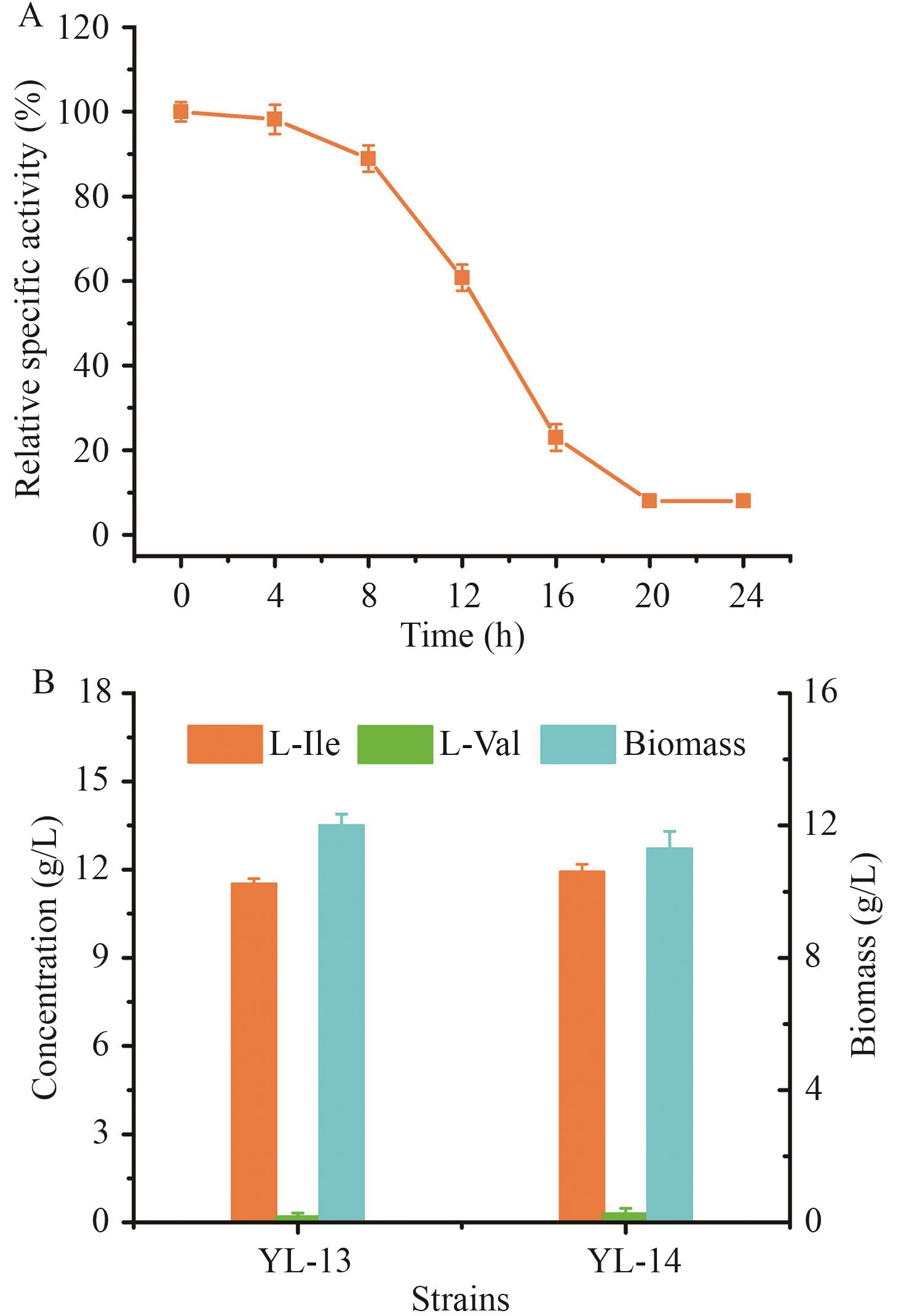
Fig. 9 Effects of activating glyoxylate cycle and dynamically attenuating TCA cycle on strains' performancesA: α-Ketoglutarate dehydrogenase activity in strain YL-14. B: Production performances of YL-13 and YL-14
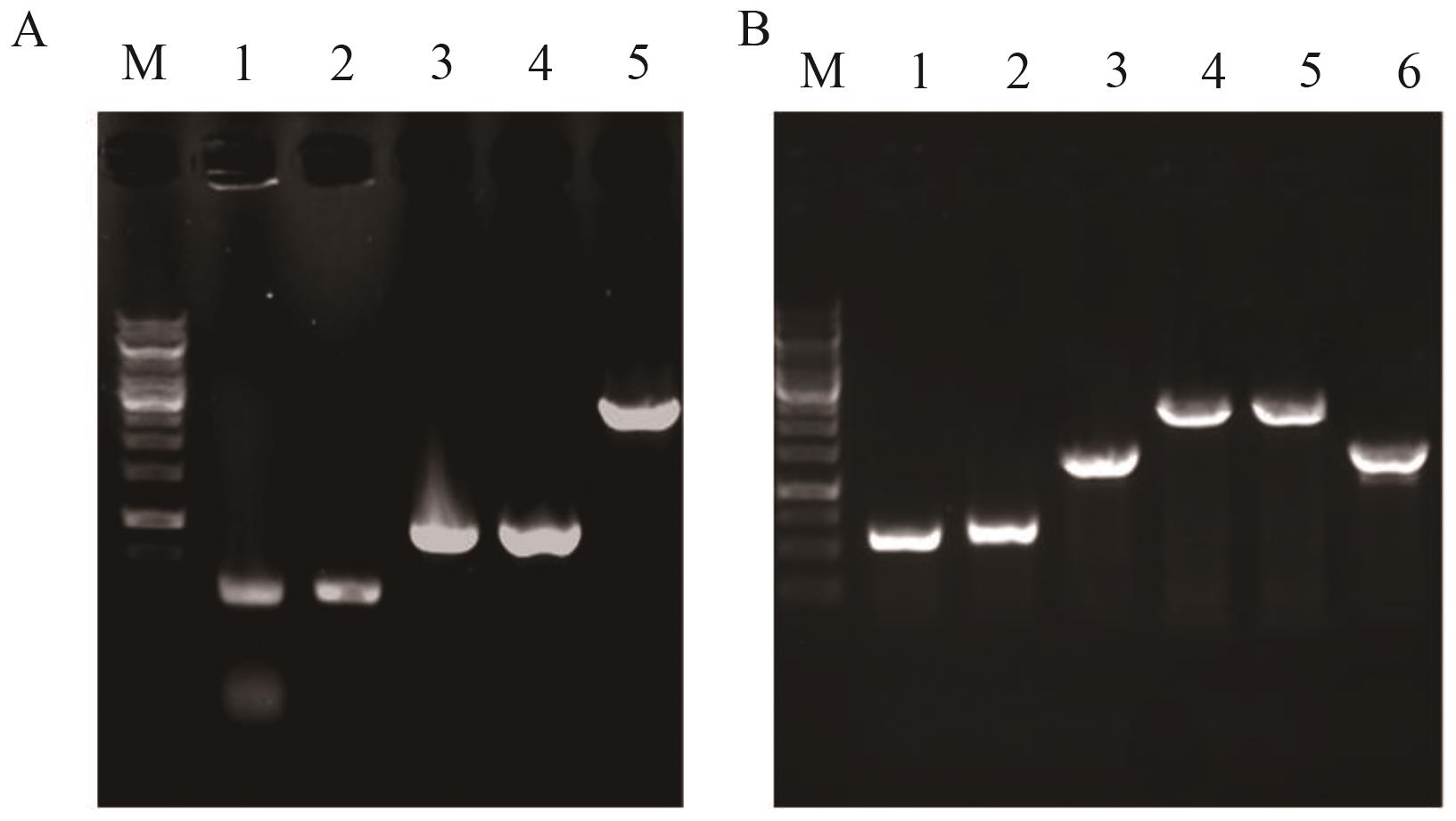
Fig. 10 Identification maps of YL-15 and YL-16A: Identification map of YL-15; M: DNA marker; 1: upstream homology arm U brnQ; 2: downstream homology arm D brnQ; 3: U brnQ -D brnQ; 4: PCR product amplified from YL-15 genomic DNA; 5: PCR product from YL-14 genomic DNA. B: Identification map of YL-16; M: DNA marker; 1: upstream homology arm U yjiV; 2: downstream homology arm D yjiV; 3: P trc -ygaZH-T trc; 4: U yjiV -P trc -ygaZH-T trc -D yjiV; 5: PCR product amplified from YL-16 genomic DNA; 6: PCR product amplified from YL-15 genomic DNA
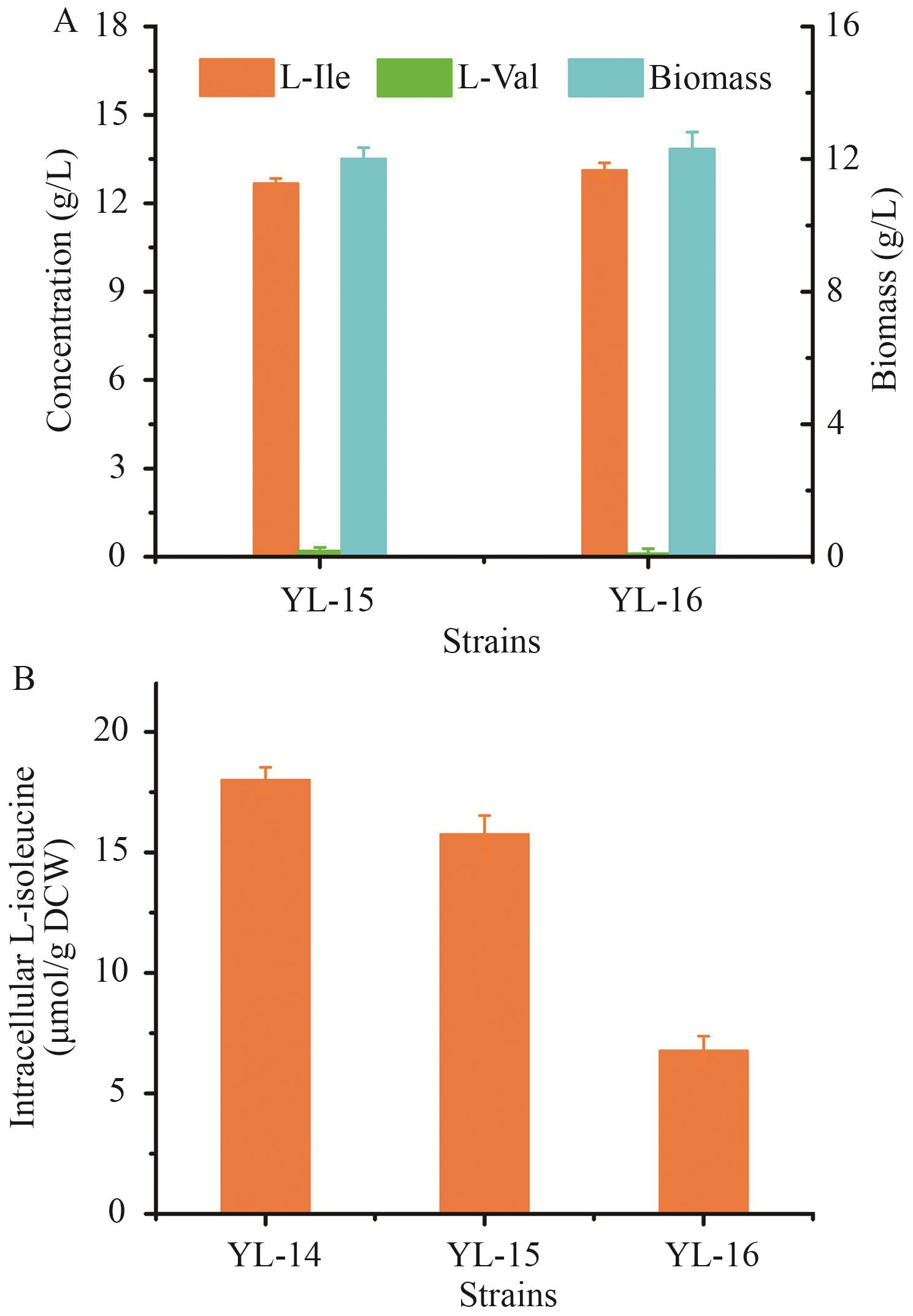
Fig. 11 Effects of modifying L-isoleucine transportation system on production performancesA: Fermatention performances of YL-15 and YL-16. B: Intracellular L-isoleucine levels in YL-14, YL-15, and YL-16
| [1] | 陈宁. 氨基酸工艺学 [M]. 2版. 北京: 中国轻工业出版社, 2020. |
| Chen N. Amino acid technology [M]. 2nd ed. Beijing: China Light Industry Press, 2020. | |
| [2] | Choi BH, Hyun S, Koo SH. The role of BCAA metabolism in metabolic health and disease [J]. Exp Mol Med, 2024, 56(7): 1552-1559. |
| [3] | 芦楠, 李宇虹, 陈宁, 等. L-异亮氨酸及其衍生物代谢工程研究进展 [J]. 食品与发酵工业, 2021, 47(9): 307-313. |
| Lu N, Li YH, Chen N, et al. Research progress on metabolic engineering of L-isoleucine and its derivatives [J]. Food and Fermentation Industries, 2021, 47(9): 307-313. | |
| [4] | Park JH, Oh JE, Lee KH, et al. Rational design of Escherichia coli for L-isoleucine production [J]. ACS Synth Biol, 2012, 1(11): 532-540. |
| [5] | Yu SZ, Zheng B, Chen ZY, et al. Metabolic engineering of Corynebacterium glutamicum for producing branched chain amino acids [J]. Microb Cell Fact, 2021, 20(1): 230. |
| [6] | Feierabend M, Renz A, Zelle E, et al. High-quality genome-scale reconstruction of Corynebacterium glutamicum ATCC 13032 [J]. Front Microbiol, 2021, 12: 750206. |
| [7] | Wang XY. Strategy for improving L-isoleucine production efficiency in Corynebacterium glutamicum [J]. Appl Microbiol Biotechnol, 2019, 103(5): 2101-2111. |
| [8] | Li HD, Xu DQ, Zhang DZ, et al. Improve L-isoleucine production in Corynebacterium glutamicum WM001 by destructing the biosynthesis of trehalose dicorynomycolate [J]. Microbiol Res, 2023, 272: 127390. |
| [9] | Morbach S, Sahm H, Eggeling L. Use of feedback-resistant threonine dehydratases of Corynebacterium glutamicum to increase carbon flux towards l-isoleucine [J]. Appl Environ Microbiol, 1995, 61(12): 4315-4320. |
| [10] | Vogt M, Krumbach K, Bang WG, et al. The contest for precursors: channelling L-isoleucine synthesis in Corynebacterium glutamicum without byproduct formation [J]. Appl Microbiol Biotechnol, 2015, 99(2): 791-800. |
| [11] | Zhang YC, Liu YD, Zhang SY, et al. Metabolic engineering of Corynebacterium glutamicum WM001 to improve l-isoleucine production [J]. Biotechnol Appl Biochem, 2021, 68(3): 568-584. |
| [12] | Hao YN, Pan XW, You JJ, et al. Microbial production of branched chain amino acids: Advances and perspectives [J]. Bioresour Technol, 2024, 397: 130502. |
| [13] | Song J, Zhuang MM, Du CY, et al. Metabolic engineering of Escherichia coli for self-induced production of L-isoleucine [J]. ACS Synth Biol, 2025, 14(1): 179-192. |
| [14] | Zhang QQ, Wang YH, Wang XL, et al. Metabolic engineering of Escherichia coli for efficient L-isoleucine production based on the citramalate pathway [J]. J Agric Food Chem, 2025, 73(19): 11900-11911. |
| [15] | Lu N, Wei MH, Yang XJ, et al. Growth-coupled production of L-isoleucine in Escherichia coli via metabolic engineering [J]. Metab Eng, 2024, 86: 181-193. |
| [16] | Shi CR, Huo XJ, You R, et al. High yield production of L-isoleucine through readjusting the ratio of two direct precursors in Escherichia coli [J]. Bioresour Technol, 2025, 418: 131889. |
| [17] | Li YF, Lin ZQ, Huang C, et al. Metabolic engineering of Escherichia coli using CRISPR-Cas9 meditated genome editing [J]. Metab Eng, 2015, 31: 13-21. |
| [18] | Zhang Y, Wei MH, Zhao GH, et al. High-level production of l-homoserine using a non-induced, non-auxotrophic Escherichia coli chassis through metabolic engineering [J]. Bioresour Technol, 2021, 327: 124814. |
| [19] | Ding MZ, Zhou X, Yuan YJ. Metabolome profiling reveals adaptive evolution of Saccharomyces cerevisiae during repeated vacuum fermentations [J]. Metabolomics, 2010, 6(1): 42-55. |
| [20] | Hao YN, Ma Q, Liu XQ, et al. High-yield production of L-valine in engineered Escherichia coli by a novel two-stage fermentation [J]. Metab Eng, 2020, 62: 198-206. |
| [21] | 靳鑫, 王苏蒙, 祁庆生, 等. 谷氨酸棒杆菌生产异亮氨酸辅因子策略及其基因组整合研究进展 [J]. 生物技术进展, 2022, 12(2): 176-188. |
| Jin X, Wang SM, Qi QS, et al. Research progress on cofactor strategy and genome integration for isoleucine production in Corynebacterium glutamicum [J]. Biotechnology Advances, 2022, 12(2), 176-188. | |
| [22] | Hirasawa T, Satoh Y, Koma D. Production of aromatic amino acids and their derivatives by Escherichia coli and Corynebacterium glutamicum [J]. World J Microbiol Biotechnol, 2025, 41(2): 65. |
| [23] | Koendjbiharie JG, van Kranenburg R, Kengen SWM. The PEP-pyruvate-oxaloacetate node: variation at the heart of metabolism [J]. FEMS Microbiol Rev, 2021, 45(3): fuaa061. |
| [24] | Klaffl S, Eikmanns BJ. Genetic and functional analysis of the soluble oxaloacetate decarboxylase from Corynebacterium glutamicum [J]. J Bacteriol, 2010, 192(10): 2604-2612. |
| [25] | Sawada K, Zen-in S, Wada M, et al. Metabolic changes in a pyruvate kinase gene deletion mutant of Corynebacterium glutamicum ATCC 13032 [J]. Metab Eng, 2010, 12(4): 401-407. |
| [26] | Mu QX, Zhang SS, Mao XJ, et al. Highly efficient production of L-homoserine in Escherichia coli by engineering a redox balance route [J]. Metab Eng, 2021, 67: 321-329. |
| [27] | Sun T, Zhao YC, Wang JJ, et al. Increasing 1, 4-diaminobutane production in Escherichia coli by optimization of cofactor PLP and NADPH synthesis [J]. Molecules, 2024, 29(13): 3094. |
| [28] | Ma WJ, Wang JL, Li Y, et al. Enhancing pentose phosphate pathway in Corynebacterium glutamicum to improve L-isoleucine production [J]. Biotechnol Appl Biochem, 2016, 63(6): 877-885. |
| [29] | Wang FA, Cai NY, Leng YL, et al. Metabolic engineering of Corynebacterium glutamicum for the high-level production of L-valine under aerobic conditions [J]. ACS Synth Biol, 2024, 13(9): 2861-2872. |
| [30] | Zhao L, Zhang WH, Wang Q, et al. A novel NADH-dependent leucine dehydrogenase for multi-step cascade synthesis of L-phosphinothricin [J]. Enzyme Microb Technol, 2023, 166: 110225. |
| [31] | Liu P, Zhang B, Yao ZH, et al. Multiplex design of the metabolic network for production of l-homoserine in Escherichia coli [J]. Appl Environ Microbiol, 2020, 86(20): e01477-20. |
| [32] | Ding ZX, Fang Y, Zhu LF, et al. Deletion of ArcA, iclR, and tdcC in Escherichia coli to improve L-threonine production [J]. Biotechnol Appl Biochem, 2019, 66(5): 794-807. |
| [33] | Wei MH, Li GR, Xie HX, et al. Sustainable production of 4-hydroxyisoleucine with minimised carbon loss by simultaneously utilising glucose and xylose in engineered Escherichia coli [J]. Bioresour Technol, 2022, 354: 127196. |
| [1] | YAN Meng-yang, LIANG Xiao-yang, DAI Jun-ang, ZHANG Yan, GUAN Tuan, ZHANG Hui, LIU Liang-bo, SUN Zhi-hua. Screening of Amoxicillin-degrading Bacteria and Study on Its Degradation Mechanisms [J]. Biotechnology Bulletin, 2025, 41(9): 314-325. |
| [2] | HUANG Xu-sheng, ZHOU Ya-li, CHAI Xu-dong, WEN Jing, WANG Ji-ping, JIA Xiao-yun, LI Run-zhi. Cloning of Plastidial PfLPAT1B Gene from Perilla frutescens and Its Functional Analysis in Oil Biosynthesis [J]. Biotechnology Bulletin, 2025, 41(7): 226-236. |
| [3] | LU Tian-yi, LI Ai-peng, FEI Qiang. Research Progress in the Biosynthesis of Polylactic Acid [J]. Biotechnology Bulletin, 2025, 41(4): 47-60. |
| [4] | RAO Jun, ZHAO Chen, LI Duan-hua, LIAO Hao, HUANG Jia-yu, WANG Lu. Application of Auto-induction Strategy in Ergothioneine Biosynthesis [J]. Biotechnology Bulletin, 2025, 41(1): 333-346. |
| [5] | ZHANG Jing-an, HU Xiao-long, CAO Bei-bei, LIAO Min, SHU Chang-long, ZHANG Jie, WANG Kui, CAO Hai-qun. Construction and Characterization of Rapid Visual Expression Vector for Bacillus thuringiensis [J]. Biotechnology Bulletin, 2025, 41(1): 95-102. |
| [6] | ZHANG Mei-yu, ZHAO Yu-bin, WANG Ling-yun, SONG Yuan-da, ZHAO Xin-he, REN Xiao-jie. Research Progress in the Production of Functional Fatty Acid DHA by Microalga Thraustochytrids [J]. Biotechnology Bulletin, 2024, 40(6): 81-94. |
| [7] | WANG Zhou, YU Jie, WANG Jin-hua, WANG Yong-ze, ZHAO Xiao. Anaerobic Expression of Lactate Dehydrogenase to Improve the D-lactic Acid Optical Purity Procluced by Escherichia coli [J]. Biotechnology Bulletin, 2024, 40(5): 290-299. |
| [8] | ZHUANG Ke, LIANG Zhi-xuan, HE Ying-ting, XIE Qiu-ling. Transfer of Antibiotic-resistance Gene AmpR by Escherichia coli DH5α Through Outer Membrane Vesicles [J]. Biotechnology Bulletin, 2024, 40(12): 275-281. |
| [9] | HE Si-cheng, ZHANG Zi-yuan, HAN Yu-qing, MIAO Lin, ZHANG Cui-ying, YU Ai-qun. Research Progress in the Production of Polyunsaturated Fatty Acids by Yarrowia lipolytica Cell Factories [J]. Biotechnology Bulletin, 2024, 40(1): 72-85. |
| [10] | LI Liang, XU Shan-shan, JIANG Yan-jun. Research Progress in the Production of Ergothioneine by Biosynthesis [J]. Biotechnology Bulletin, 2024, 40(1): 86-99. |
| [11] | YANG Hong-yan, HAN Xiao, YANG Jian-jun. Scaling up Production of pDNA Plasmids in Disposable Bioreactors [J]. Biotechnology Bulletin, 2024, 40(1): 168-175. |
| [12] | XUE Ning, WANG Jin, LI Shi-xin, LIU Ye, CHENG Hai-jiao, ZHANG Yue, MAO Yu-feng, WANG Meng. Construction of L-phenylalanine High-producing Corynebacterium glutamicum Engineered Strains via Multi-gene Simultaneous Regulation Combined with High-throughput Screening [J]. Biotechnology Bulletin, 2023, 39(9): 268-280. |
| [13] | CHENG Ya-nan, ZHANG Wen-cong, ZHOU Yuan, SUN Xue, LI Yu, LI Qing-gang. Synthetic Pathway Construction of Producing 2'-fucosyllactose by Lactococcus lactis and Optimization of Fermentation Medium [J]. Biotechnology Bulletin, 2023, 39(9): 84-96. |
| [14] | ZHAO Si-jia, WANG Xiao-lu, SUN Ji-lu, TIAN Jian, ZHANG Jie. Modification of Pichia pastoris for Erythritol Production by Metabolic Engineering [J]. Biotechnology Bulletin, 2023, 39(8): 137-147. |
| [15] | LI Yu-zhen, MEI Tian-xiu, LI Zhi-wen, WANG Qi, LI Jun, ZOU Yue, ZHAO Xin-qing. Advances in Genomic Studies and Metabolic Engineering of Red Yeasts [J]. Biotechnology Bulletin, 2023, 39(7): 67-79. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||