








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (12): 254-266.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0469
Previous Articles Next Articles
ZENG Liang-qin1,2( ), DONG Chen-wen-hua1, LIN Chun1, LIU Zheng-jie1, MAO Zi-chao1(
), DONG Chen-wen-hua1, LIN Chun1, LIU Zheng-jie1, MAO Zi-chao1( )
)
Received:2025-05-08
Online:2025-12-26
Published:2026-01-06
Contact:
MAO Zi-chao
E-mail:zengliangqin2007@163.com;zmao@ynau.edu.cn
ZENG Liang-qin, DONG Chen-wen-hua, LIN Chun, LIU Zheng-jie, MAO Zi-chao. Identification of the AsparagusCYP51 Gene Family and the Response to Abiotic Stress[J]. Biotechnology Bulletin, 2025, 41(12): 254-266.
| 引物编号 Primer number | 引物序列 Primer sequence(5'-3') | 目的 Purpose |
|---|---|---|
| AtaCYP51G2-1F | ATGGATTTAACAGAGAACAAGTTCTTG | 基因克隆 |
| AtaCYP51G2-1R | TTATTCAACCGAAAGCTTCCTCC | 基因克隆 |
| AtaCYP51G2-2F | TGGTCGACGTACTAGATGGATTTAACAGAGAACAAGTTCTTGAGCA | 载体构建 |
| AtaCYP51G2-2R | GCTCACCATCACTAGTTCAACCGAAAGCTTCCTCCTCTTGT | 载体构建 |
| AtaCYP51G2-qF | TCCCGTAAATGCTCAGGTCG | RT-qPCR |
| AtaCYP51G2-qR | GTTGAGGTGATCGAGCTGGT | RT-qPCR |
| ACTIN-qF | TGACTACGAGCAGGAGATGGAA | RT-qPCR |
| ACTIN-qR | AAACGAGGGCTGGAACAAGA | RT-qPCR |
Table 1 Primers used in this study
| 引物编号 Primer number | 引物序列 Primer sequence(5'-3') | 目的 Purpose |
|---|---|---|
| AtaCYP51G2-1F | ATGGATTTAACAGAGAACAAGTTCTTG | 基因克隆 |
| AtaCYP51G2-1R | TTATTCAACCGAAAGCTTCCTCC | 基因克隆 |
| AtaCYP51G2-2F | TGGTCGACGTACTAGATGGATTTAACAGAGAACAAGTTCTTGAGCA | 载体构建 |
| AtaCYP51G2-2R | GCTCACCATCACTAGTTCAACCGAAAGCTTCCTCCTCTTGT | 载体构建 |
| AtaCYP51G2-qF | TCCCGTAAATGCTCAGGTCG | RT-qPCR |
| AtaCYP51G2-qR | GTTGAGGTGATCGAGCTGGT | RT-qPCR |
| ACTIN-qF | TGACTACGAGCAGGAGATGGAA | RT-qPCR |
| ACTIN-qR | AAACGAGGGCTGGAACAAGA | RT-qPCR |
物种 Species | 基因名称 Gene name | 基因编号 Gene ID | 蛋白长度 Protein length (aa) | 分子量 Molecular weight (kD) | 等电点 Isoelectric point | 脂肪指数 Aliphatic index | 平均亲水性 Grand average of hydropathicity | 不稳定指数 Instability index | 亚细胞定位预测 Predict subcellular localization |
|---|---|---|---|---|---|---|---|---|---|
大理天门冬 A. taliensis | AtaCYP51G1 | Ata0G001960.1 | 488 | 55.4 | 8.7 | 93.3 | -0.116 | 35.7 | 质膜 Plasma membrane |
| AtaCYP51G2 | Ata09G008170.1 | 488 | 55.4 | 8.69 | 94.1 | -0.101 | 34.05 | 内膜 Inner membrane | |
芦笋 A. officinalis | AoCYP51G1 | AsparagusV1_Unassigned.240 | 510 | 58.0 | 8.94 | 93.84 | -0.106 | 34.85 | 质膜 Plasma membrane |
文竹 A. setaceus | AsCYP51G1 | Ase07G1939-RA | 488 | 55.5 | 8.55 | 91.68 | -0.14 | 36.22 | 质膜 Plasma membrane |
Table 2 Physicochemical characteristics of CYP51 gene family in Asparagus species
物种 Species | 基因名称 Gene name | 基因编号 Gene ID | 蛋白长度 Protein length (aa) | 分子量 Molecular weight (kD) | 等电点 Isoelectric point | 脂肪指数 Aliphatic index | 平均亲水性 Grand average of hydropathicity | 不稳定指数 Instability index | 亚细胞定位预测 Predict subcellular localization |
|---|---|---|---|---|---|---|---|---|---|
大理天门冬 A. taliensis | AtaCYP51G1 | Ata0G001960.1 | 488 | 55.4 | 8.7 | 93.3 | -0.116 | 35.7 | 质膜 Plasma membrane |
| AtaCYP51G2 | Ata09G008170.1 | 488 | 55.4 | 8.69 | 94.1 | -0.101 | 34.05 | 内膜 Inner membrane | |
芦笋 A. officinalis | AoCYP51G1 | AsparagusV1_Unassigned.240 | 510 | 58.0 | 8.94 | 93.84 | -0.106 | 34.85 | 质膜 Plasma membrane |
文竹 A. setaceus | AsCYP51G1 | Ase07G1939-RA | 488 | 55.5 | 8.55 | 91.68 | -0.14 | 36.22 | 质膜 Plasma membrane |
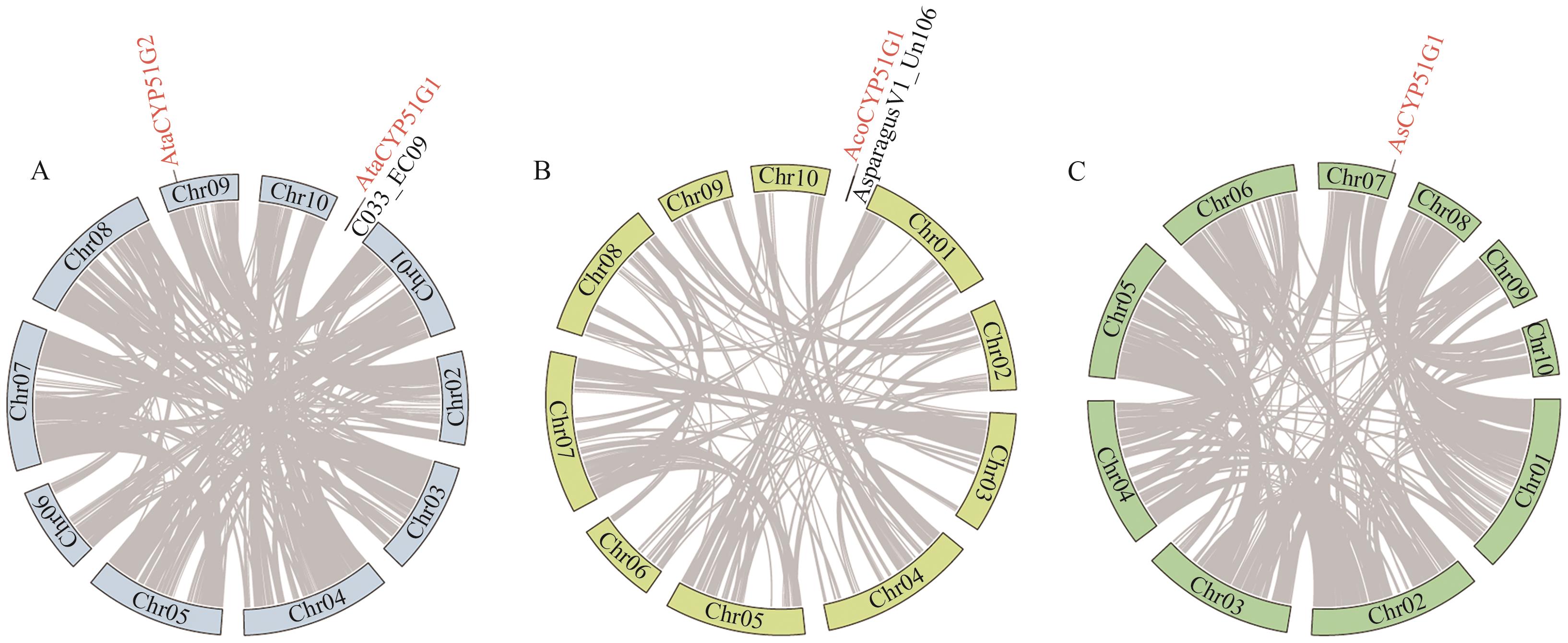
Fig. 1 Chromosomal localization of the CYP51 gene family in Asparagus speciesA-C indicate the chromosomal localization of the CYP51 gene family in A. taliensis, A. officinalis, and A. setaceus, respectively. The CYP51 genes in Asparagus species are marked in red. Gray lines indicate intraspecific collinear gene pairs
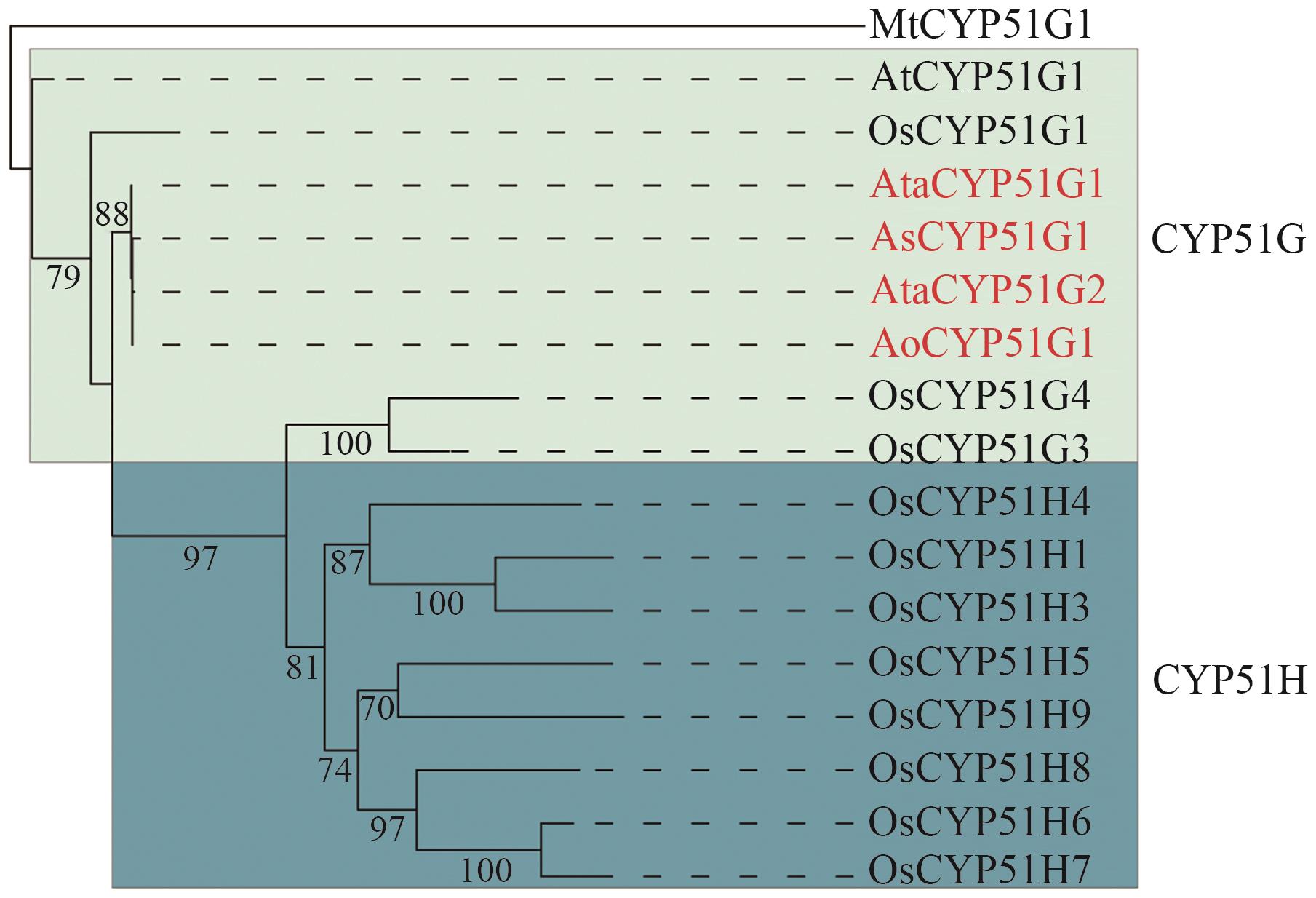
Fig. 2 Phylogenetic tree of CYP51 proteinsThe CYP51 genes in Asparagus species are marked in red. Ata: A. taliensis; Ao: A. officinalis; As: A. setaceus; At: A. thaliana; Os: O. sativa; Mt: Mycobacterium tuberculosis
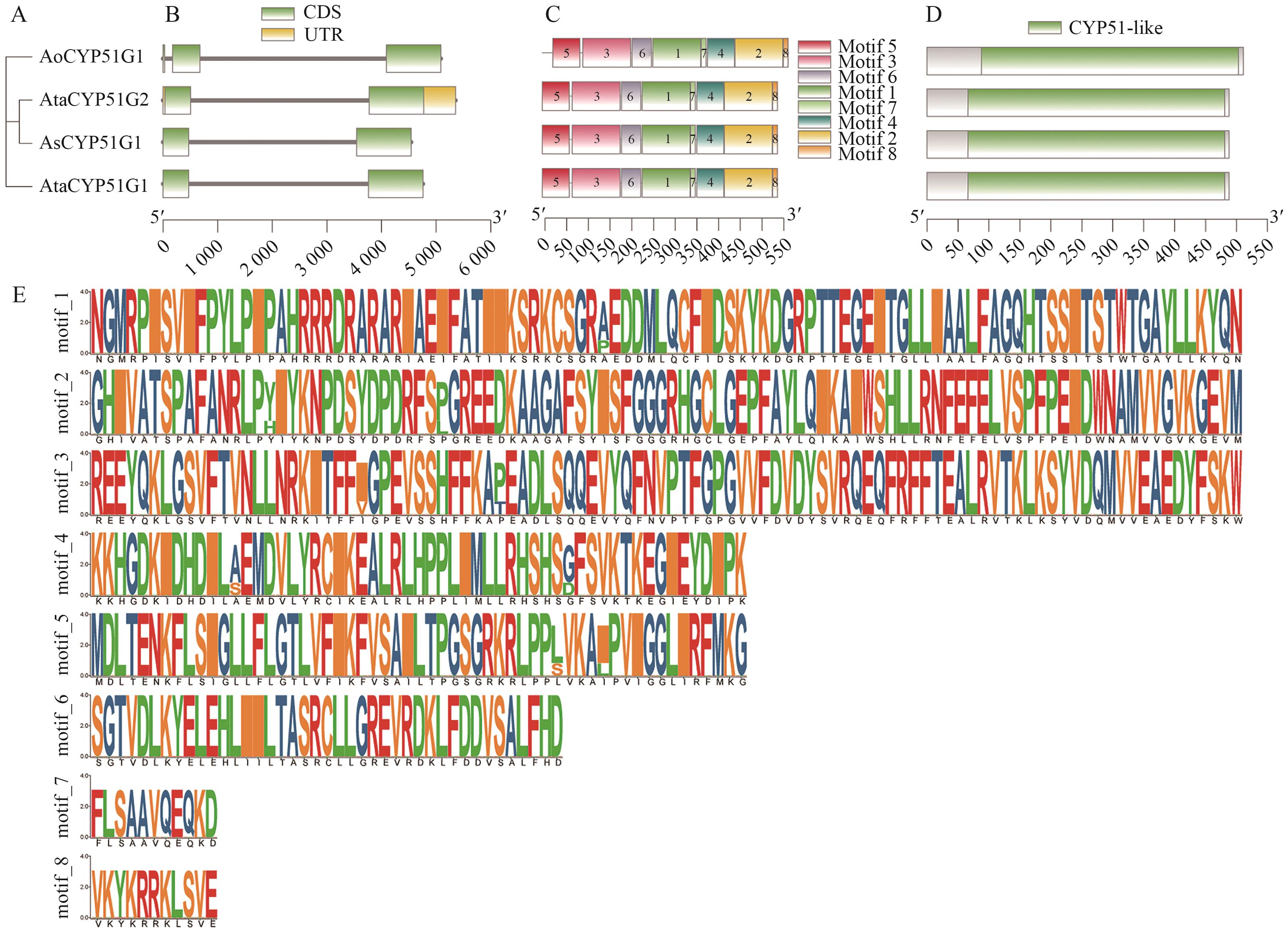
Fig. 3 Gene structure, conserved motifs, and conserved domain analysis of the CYP51 gene family in Asparagus speciesA: Phylogenetic tree of Asparagus CYP51 protein. B: Gene structure. C: Conserved motifs. D: Conserved domains. E: Conserved motif annotations
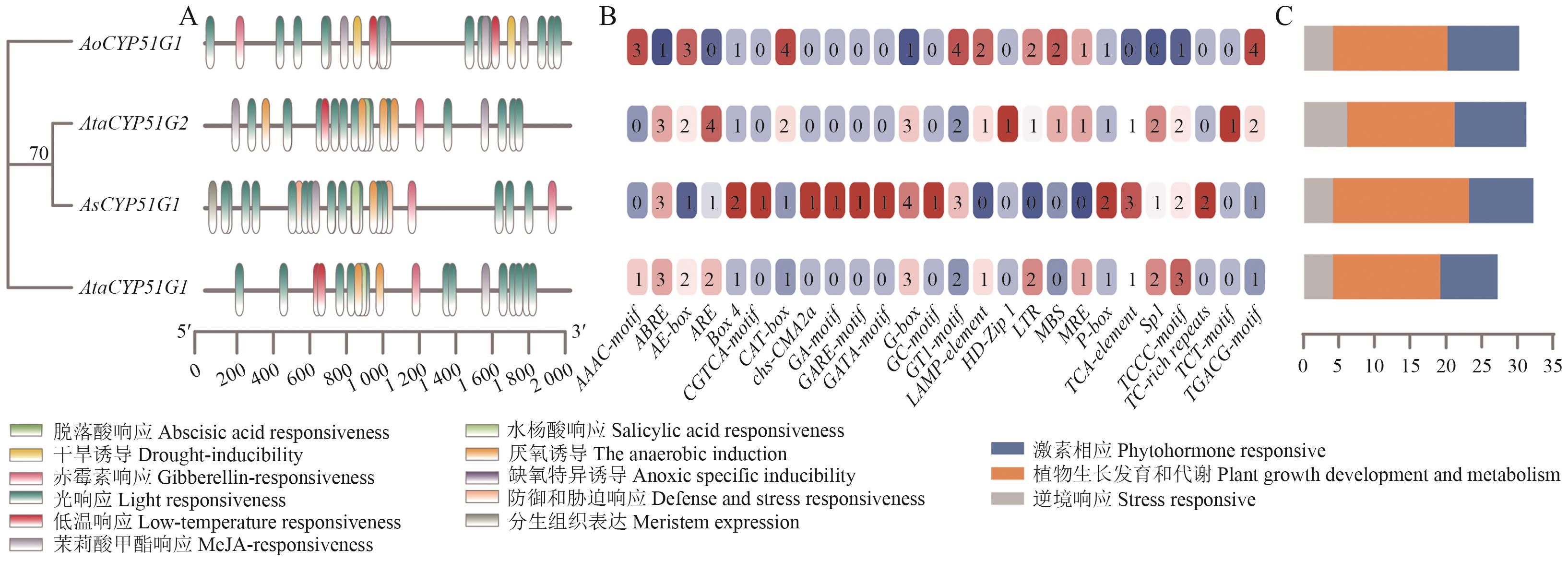
Fig. 4 Cis-acting element analysis of the CYP51 gene family in Asparagus speciesA: Distribution of cis-acting elements. B: Number of cis-acting elements. C: Types and quantities of cis-acting elements
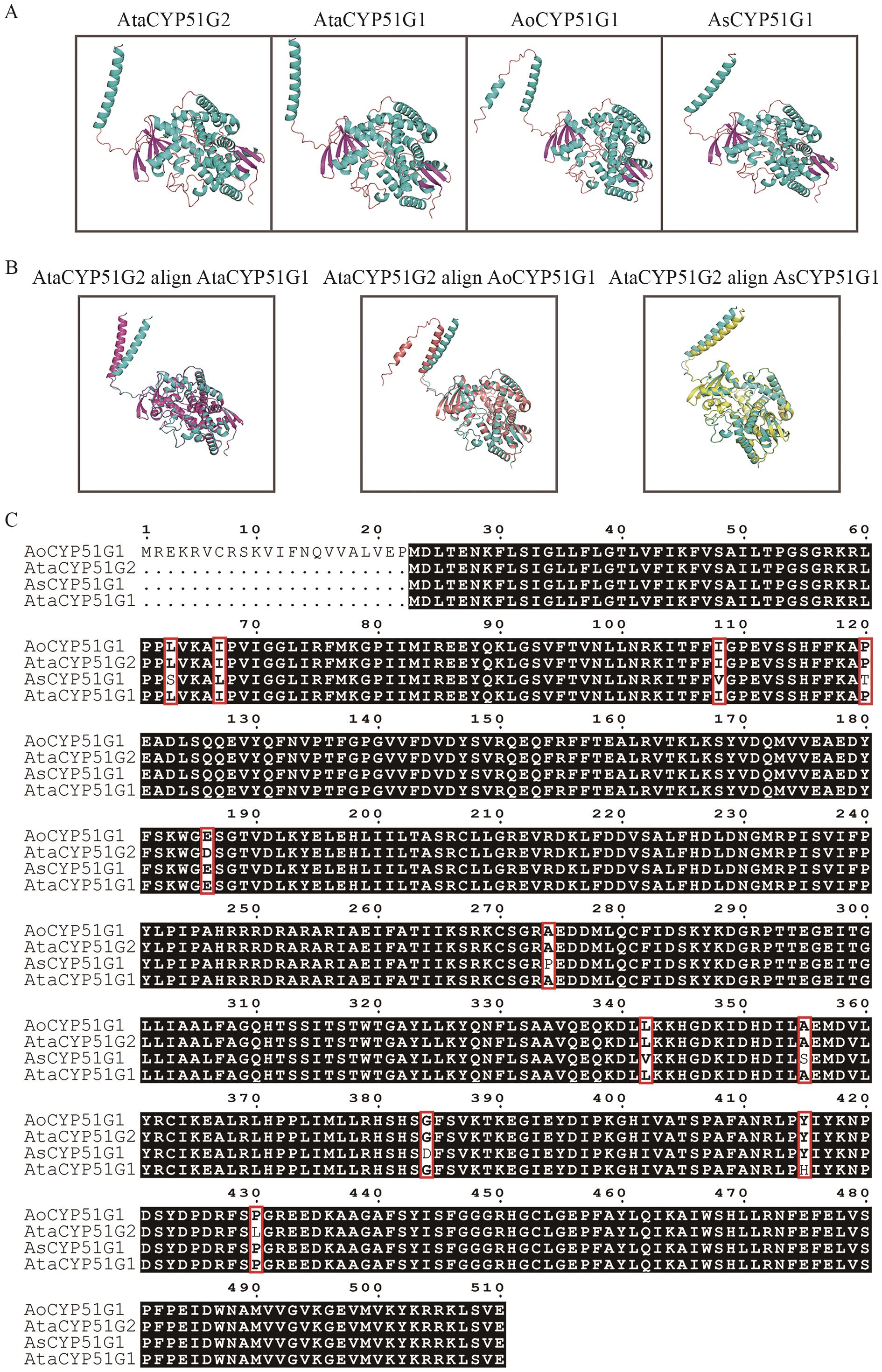
Fig. 5 Primary sequence alignment, tertiary structure prediction and alignment of CYP51 proteins in Asparagus speciesA: Predicting tertiary structure of proteins. B: Alignment of tertiary structures. C: Multiple-sequence alignment of primary structures
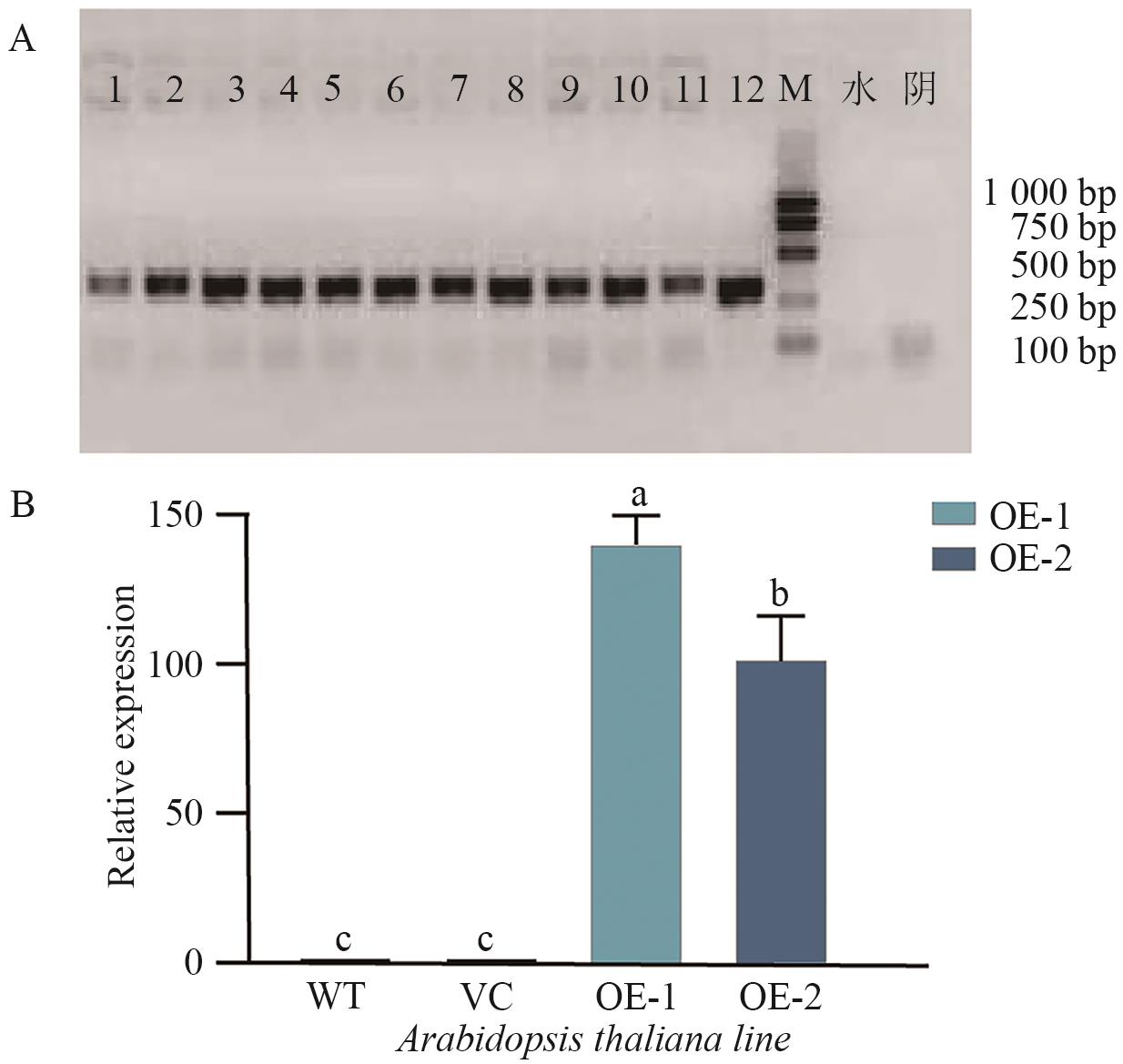
Fig. 7 Identification of AtaCYP51G2 transgenic Arabidopsis linesA: Genomic PCR analysis of T2 plants. Lane 1-6: line OE-1; lane 7-12: line OE-2. B: Relative expression of AtaCYP51G2 in wild type (WT), vector control (VC), and transgenic lines (OE-1 and OE-2), determined by qRT-PCR. Data are presented as mean±SD (n=3). Different letters indicate statistically significant differences (P<0.05)
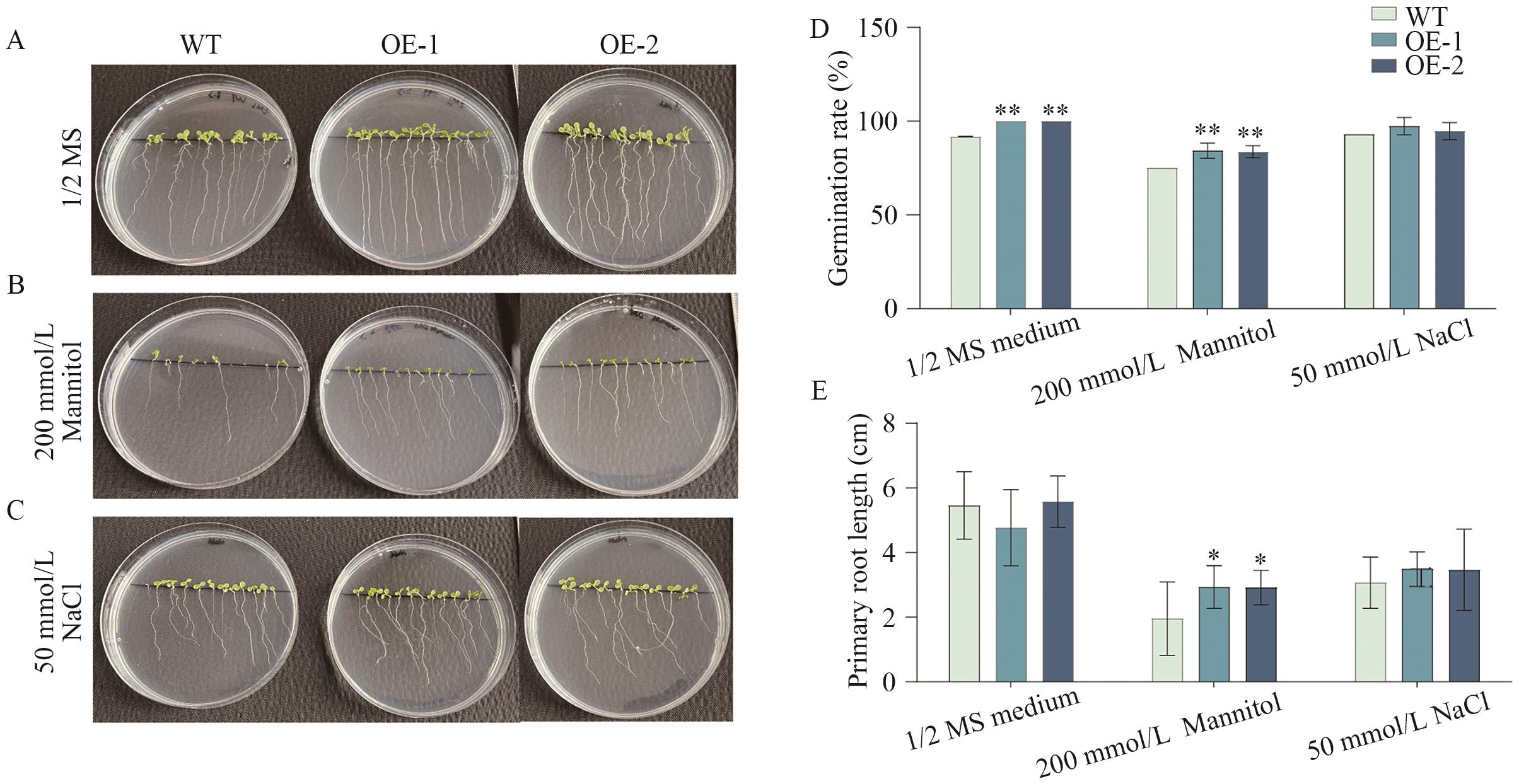
Fig. 9 Evaluation of seed germination and root length in transgenic Arabidopsis under salt and osmotic stressA-C: Phenotypic observation of Arabidopsis grown on solid medium. D: Measurement of Arabidopsis seed germination rate. E: Measurement of Arabidopsis primary root length
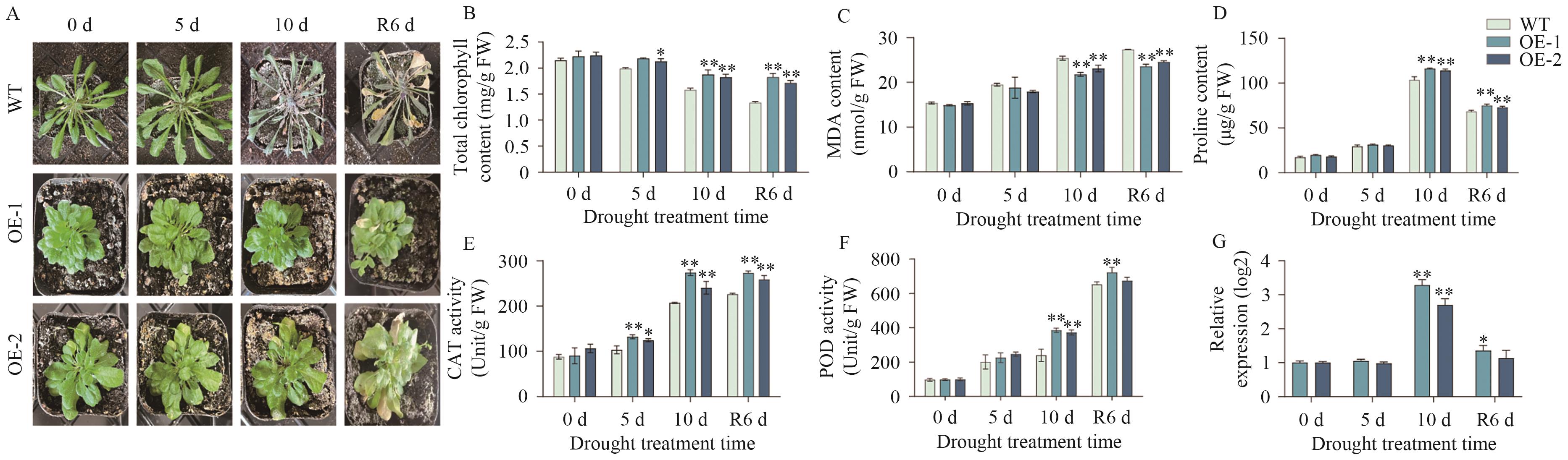
Fig. 10 Phenotype and physiological and biochemical measurements of AtaCYP51G2-overexpressing Arabidopsis under drought stressA: Phenotype of wild-type (WT) and AtaCYP51G2-overexpressing (OE) Arabidopsis lines under drought stress; B-F: the physiological and biochemical indicators of WT and OE Arabidopsis plants under drought stress, respectively: total chlorophyll content, malondialdehyde content, proline content, catalase activity, and peroxidase activity. The same below. G: Relative gene expressions
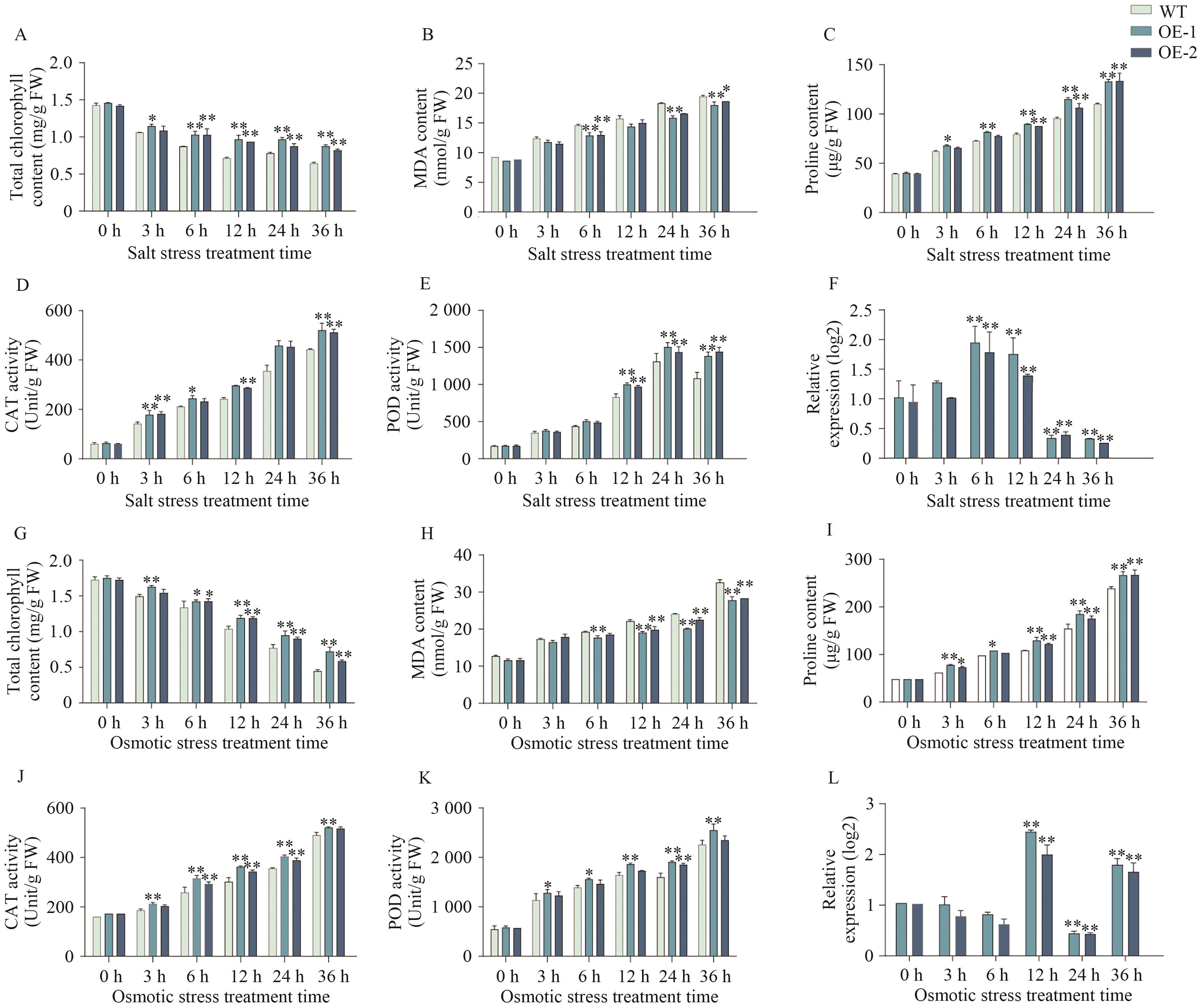
Fig. 11 Physiological measurements of AtaCYP51G2-overexpressing Arabidopsis under salt and osmotic stressA-F: Physiological and biochemical parameters and the relative transcript levels of AtaCYP51G2 overexpressing lines under salt stress, whereas panel G-L display the corresponding data under osmotic stress
| [1] | Zhu JK. Salt and drought stress signal transduction in plants [J]. Annu Rev Plant Biol, 2002, 53: 247-273. |
| [2] | Hartmann MA. Plant sterols and the membrane environment [J]. Trends Plant Sci, 1998, 3(5): 170-175. |
| [3] | Lepesheva GI, Waterman MR. Sterol 14α-demethylase cytochrome P450 (CYP51), a P450 in all biological Kingdoms [J]. Biochim Biophys Acta BBA Gen Subj, 2007, 1770(3): 467-477. |
| [4] | Du YL, Fu XZ, Chu YY, et al. Biosynthesis and the roles of plant sterols in development and stress responses [J]. Int J Mol Sci, 2022, 23(4): 2332. |
| [5] | Jiao ZL, Yin LJ, Zhang QM, et al. The putative obtusifoliol 14α-demethylase OsCYP51H3 affects multiple aspects of rice growth and development [J]. Physiol Plant, 2022, 174(5): e13764. |
| [6] | Xia KF, Ou XJ, Tang HD, et al. Rice microRNA Osa-miR1848 targets the obtusifoliol 14α-demethylase gene OsCYP51G3 and mediates the biosynthesis of phytosterols and brassinosteroids during development and in response to stress [J]. New Phytol, 2015, 208(3): 790-802. |
| [7] | 唐万虎, 张祖新, 邹锡玲, 等. 玉米耐渍功能基因组分析及相关基因Sicyp51的鉴定与克隆 [J]. 中国科学C辑: 生命科学, 2005, 35(1): 29-36. |
| Tang WH, Zhang ZX, Zou XL, et al. Genome analysis of waterlogging tolerance function in maize and identification and cloning of related gene Sicyp51 [J]. Sci China Ser C, 2005, 35(1): 29-36. | |
| [8] | 温晶媛, 李颖, 丁声颂, 等. 中国百合科天门冬属九种药用植物的药理作用筛选 [J]. 上海医科大学学报, 1993, 20(2): 107-111. |
| Wen JY, Li Y, Ding SS, et al. Pharmacological screening of 9 medicinal plants of the genus Asparagus (Liliaceae) in China [J]. Fudan Univ J Med Sci, 1993, 20(2): 107-111. | |
| [9] | Zeng LQ, Brown SE, Wu H, et al. Comprehensive genome-wide analysis of the HMGR gene family of Asparagus taliensis and functional validation of AtaHMGR10 under different abiotic stresses [J]. Front Plant Sci, 2025, 16: 1455592. |
| [10] | Zhang XR, Henriques R, Lin SS, et al. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method [J]. Nat Protoc, 2006, 1(2): 641-646. |
| [11] | Paquette SM, Jensen K, Bak S. A web-based resource for the Arabidopsis P450, cytochromes b5, NADPH-cytochrome P450 reductases, and family 1 glycosyltransferases (http://www. P450. kvl. dk) [J]. Phytochemistry, 2009, 70(17-18): 1940-1947. |
| [12] | Malhotra K, Franke J. Cytochrome P450 monooxygenase-mediated tailoring of triterpenoids and steroids in plants [J]. Beilstein J Org Chem, 2022, 18: 1289-1310. |
| [13] | McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools [J]. Nucleic Acids Res, 2004, 32(Web Server issue): W20-W25. |
| [14] | Chen CJ, Wu Y, Li JW, et al. TBtools-II: a “one for all, all for one” bioinformatics platform for biological big-data mining [J]. Mol Plant, 2023, 16(11): 1733-1742. |
| [15] | Lu SN, Wang JY, Chitsaz F, et al. CDD/SPARCLE: the conserved domain database in 2020 [J]. Nucleic Acids Res, 2020, 48(D1): D265-D268. |
| [16] | Yu CS, Chen YC, Lu CH, et al. Prediction of protein subcellular localization [J]. Proteins, 2006, 64(3): 643-651. |
| [17] | Wang YP, Li JP, Paterson AH. MCScanX-transposed: detecting transposed gene duplications based on multiple colinearity scans [J]. Bioinformatics, 2013, 29(11): 1458-1460. |
| [18] | Inagaki YS, Etherington G, Geisler K, et al. Investigation of the potential for triterpene synthesis in rice through genome mining and metabolic engineering [J]. New Phytol, 2011, 191(2): 432-448. |
| [19] | Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput [J]. Nucleic Acids Res, 2004, 32(5): 1792-1797. |
| [20] | Bailey TL, Johnson J, Grant CE, et al. The MEME suite [J]. Nucleic Acids Res, 2015, 43(w1): W39-W49. |
| [21] | Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server [J]. Nucleic Acids Res, 2014, 42(Web Server issue): W320-W324. |
| [22] | Callaway E. AI protein-prediction tool AlphaFold3 is now more open [J]. Nature, 2024, 635(8039): 531-532. |
| [23] | Abramson J, Adler J, Dunger J, et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3 [J]. Nature, 2024, 630(8016): 493-500. |
| [24] | Bowie JU, Lüthy R, Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure [J]. Science, 1991, 253(5016): 164-170. |
| [25] | Lüthy R, Bowie JU, Eisenberg D. Assessment of protein models with three-dimensional profiles [J]. Nature, 1992, 356(6364): 83-85. |
| [26] | Colovos C, Yeates TO. Verification of protein structures: patterns of nonbonded atomic interactions [J]. Protein Sci, 1993, 2(9): 1511-1519. |
| [27] | Pontius J, Richelle J, Wodak SJ. Deviations from standard atomic volumes as a quality measure for protein crystal structures [J]. J Mol Biol, 1996, 264(1): 121-136. |
| [28] | DeLano WL. Pymol: An open-source molecular graphics tool [J]. CCP4 Newsl Protein Crystallogr, 2002, 40(1): 82-92. |
| [29] | Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements [J]. Nat Methods, 2015, 12(4): 357-360. |
| [30] | Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features [J]. Bioinformatics, 2014, 30(7): 923-930. |
| [31] | Zhang W, Li HX, Li QH, et al. Genome-wide identification, comparative analysis and functional roles in flavonoid biosynthesis of cytochrome P450 superfamily in pear (Pyrus spp.) [J]. BMC Genom Data, 2023, 24(1): 58. |
| [32] | 杨娇艳, 廖明军, 杨劭. 甾醇14α-去甲基化酶(CYP51)的研究进展 [J]. 生物工程学报, 2008, 24(10): 1681-1688. |
| Yang JY, Liao MJ, Yang S. Advances in sterol 14α-demethylase (CYP51) [J]. Chin J Biotechnol, 2008, 24(10): 1681-1688. | |
| [33] | Li SF, Wang J, Dong R, et al. Chromosome-level genome assembly, annotation and evolutionary analysis of the ornamental plant Asparagus setaceus [J]. Hortic Res, 2020, 7(1): 48. |
| [34] | Shang LG, He WC, Wang TY, et al. A complete assembly of the rice Nipponbare reference genome [J]. Mol Plant, 2023, 16(8): 1232-1236. |
| [35] | Kim HB, Schaller H, Goh CH, et al. Arabidopsis cyp51 mutant shows postembryonic seedling lethality associated with lack of membrane integrity [J]. Plant Physiol, 2005, 138(4): 2033-2047. |
| [1] | CHENG Ting-ting, LIU Jun, WANG Li-li, LIAN Cong-long, WEI Wen-jun, GUO Hui, WU Yao-lin, YANG Jing-fan, LAN Jin-xu, CHEN Sui-qing. Genome-wide Identification of the Chalcone Isomerase Gene Family in Eucommia ulmoides and Analysis of Their Expression Patterns [J]. Biotechnology Bulletin, 2025, 41(9): 242-255. |
| [2] | LI Shan, MA Deng-hui, MA Hong-yi, YAO Wen-kong, YIN Xiao. Identification and Expression Analysis of SKP1 Gene Family in Grapevine (Vitis vinifera L.) [J]. Biotechnology Bulletin, 2025, 41(9): 147-158. |
| [3] | HUANG Guo-dong, DENG Yu-xing, CHENG Hong-wei, DAN Yan-nan, ZHOU Hui-wen, WU Lan-hua. Genome-wide Identification and Expression Analysis of the ZIP Gene Family in Soybean [J]. Biotechnology Bulletin, 2025, 41(9): 71-81. |
| [4] | HUA Wen-ping, LIU Fei, HAO Jia-xin, CHEN Chen. Identification and Expression Patterns Analysis of ADH Gene Family in Salvia miltiorrhiza [J]. Biotechnology Bulletin, 2025, 41(8): 211-219. |
| [5] | LA Gui-xiao, ZHAO Yu-long, DAI Dan-dan, YU Yong-liang, GUO Hong-xia, SHI Gui-xia, JIA Hui, YANG Tie-gang. Identification of Plasma Membrane H+-ATPase Gene Family in Safflower and Expression Analysis in Response to Low Nitrogen and Low Phosphorus Stress [J]. Biotechnology Bulletin, 2025, 41(8): 220-233. |
| [6] | HUANG Shi-yu, TIAN Shan-shan, YANG Tian-wei, GAO Man-rong, ZHANG Shang-wen. Genome-wide Identification and Expression Pattern Analysis of WRI1 Gene Family in Erythropalum scandens [J]. Biotechnology Bulletin, 2025, 41(8): 242-254. |
| [7] | CHENG Xue, FU Ying, CHAI Xiao-jiao, WANG Hong-yan, DENG Xin. Identification of LHC Gene Family in Setaria italica and Expression Analysis under Abiotic Stresses [J]. Biotechnology Bulletin, 2025, 41(8): 102-114. |
| [8] | REN Rui-bin, SI Er-jing, WAN Guang-you, WANG Jun-cheng, YAO Li-rong, ZHANG Hong, MA Xiao-le, LI Bao-chun, WANG Hua-jun, MENG Ya-xiong. Identification and Expression Analysis of GH17 Gene Family of Pyrenophora graminea [J]. Biotechnology Bulletin, 2025, 41(8): 146-154. |
| [9] | ZHAI Ying, JI Jun-jie, CHEN Jiong-xin, YU Hai-wei, LI Shan-shan, ZHAO Yan, MA Tian-yi. Heterologous Overexpression of Soybean GmNF-YB24 Improves the Resistance of Transgenic Tobacco to Drought [J]. Biotechnology Bulletin, 2025, 41(8): 137-145. |
| [10] | HAN Yi, HOU Chang-lin, TANG Lu, SUN Lu, XIE Xiao-dong, LIANG Chen, CHEN Xiao-qiang. Cloning and Preliminary Functional Analysis of HvERECTA Gene in Hordeum vulgare [J]. Biotechnology Bulletin, 2025, 41(7): 106-116. |
| [11] | LI Xia, ZHANG Ze-wei, LIU Ze-jun, WANG Nan, GUO Jiang-bo, XIN Cui-hua, ZHANG Tong, JIAN Lei. Cloning and Functional Study of Transcription Factor StMYB96 in Potato [J]. Biotechnology Bulletin, 2025, 41(7): 181-192. |
| [12] | GONG Yu-han, CHEN Lan, SHANGFANG Hui-zi, HAO Ling-ying, LIU Shuo-qian. Identification and Expression Profile Analysis of the TRB Gene Family in Tea Plant [J]. Biotechnology Bulletin, 2025, 41(7): 214-225. |
| [13] | WEI Yu-jia, LI Yan, KANG Yu-han, GONG Xiao-nan, DU Min, TU Lan, SHI Peng, YU Zi-han, SUN Yan, ZHANG Kun. Cloning and Expression Analysis of the CrMYB4 Gene in Carex rigescens [J]. Biotechnology Bulletin, 2025, 41(7): 248-260. |
| [14] | ZHANG Ze, YANG Xiu-li, NING Dong-xian. Identification of 4CL Gene Family in Arachis hypogaea L. and Expression Analysis in Response to Drought and Salt Stress [J]. Biotechnology Bulletin, 2025, 41(7): 117-127. |
| [15] | ZHANG Xue-qiong, PAN Su-jun, LI Wei, DAI Liang-ying. Research Progress of Plant Phosphate Transporters in the Response to Stress [J]. Biotechnology Bulletin, 2025, 41(7): 28-36. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||