








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (11): 212-220.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0527
SANG Shi-bo( ), LI Li, ZHANG Feng-yuan, SUN Lang, SHEN Neng-shuang, CHENG Cong, REN Yan-ping, MA Li, ZHANG Hua(
), LI Li, ZHANG Feng-yuan, SUN Lang, SHEN Neng-shuang, CHENG Cong, REN Yan-ping, MA Li, ZHANG Hua( )
)
Received:2025-05-22
Online:2025-11-26
Published:2025-12-09
Contact:
ZHANG Hua
E-mail:927810653@qq.com;hazelzhang@163.com
SANG Shi-bo, LI Li, ZHANG Feng-yuan, SUN Lang, SHEN Neng-shuang, CHENG Cong, REN Yan-ping, MA Li, ZHANG Hua. Construction and Validation of the TRV-mediated Gene Silencing System in Haloxylon ammodendron[J]. Biotechnology Bulletin, 2025, 41(11): 212-220.
引物名称 Name of primer | 引物序列 Sequence of primer (5′-3′) | 用途 Application |
|---|---|---|
| pTRV2-HaCLA1-F | GTGAGTAAGGTTACCGAATTCAGGAAGACCTCAGGGCTAGCA | 沉默载体构建 Silent vector construction |
| pTRV2-HaCLA1-R | GAGACGCGTGAGCTCGGTACCCGTCCACTGGACCAATGTAGTACA | |
| pTRV2-HaNAC3-F | TGAGTAAGGTTACCGAATTCGTACTACGAGGGAAAATATCCATCATC | |
| pTRV2-HaNAC3-R | GAGACGCGTGAGCTCGGTACCTCATCTTGATCATCATTGTCTTCCTC | |
| 18SrRNA-F | CTCTGCCGTTGCTCTGATGAT | 基因相对表达量分析 Analysis of relative gene expressions |
| 18SrRNA-R | CCTTGGATGTGGTAGCCGTTC | |
| RT-qPCR-HaNAC3-F | ATCAAAACAGCCAGATGTCA | |
| RT-qPCR-HaNAC3-R | GAAGAAAGCTCCACTTCGTTG | |
| RT-qPCR-HACLA1-F | CTTTAGGGAAGCTCCAAGCATT | |
| RT-qPCR-HACLA1-R | CGCAACCTTGTGTGTTTGTCC |
Table 1 Primers used in the study
引物名称 Name of primer | 引物序列 Sequence of primer (5′-3′) | 用途 Application |
|---|---|---|
| pTRV2-HaCLA1-F | GTGAGTAAGGTTACCGAATTCAGGAAGACCTCAGGGCTAGCA | 沉默载体构建 Silent vector construction |
| pTRV2-HaCLA1-R | GAGACGCGTGAGCTCGGTACCCGTCCACTGGACCAATGTAGTACA | |
| pTRV2-HaNAC3-F | TGAGTAAGGTTACCGAATTCGTACTACGAGGGAAAATATCCATCATC | |
| pTRV2-HaNAC3-R | GAGACGCGTGAGCTCGGTACCTCATCTTGATCATCATTGTCTTCCTC | |
| 18SrRNA-F | CTCTGCCGTTGCTCTGATGAT | 基因相对表达量分析 Analysis of relative gene expressions |
| 18SrRNA-R | CCTTGGATGTGGTAGCCGTTC | |
| RT-qPCR-HaNAC3-F | ATCAAAACAGCCAGATGTCA | |
| RT-qPCR-HaNAC3-R | GAAGAAAGCTCCACTTCGTTG | |
| RT-qPCR-HACLA1-F | CTTTAGGGAAGCTCCAAGCATT | |
| RT-qPCR-HACLA1-R | CGCAACCTTGTGTGTTTGTCC |
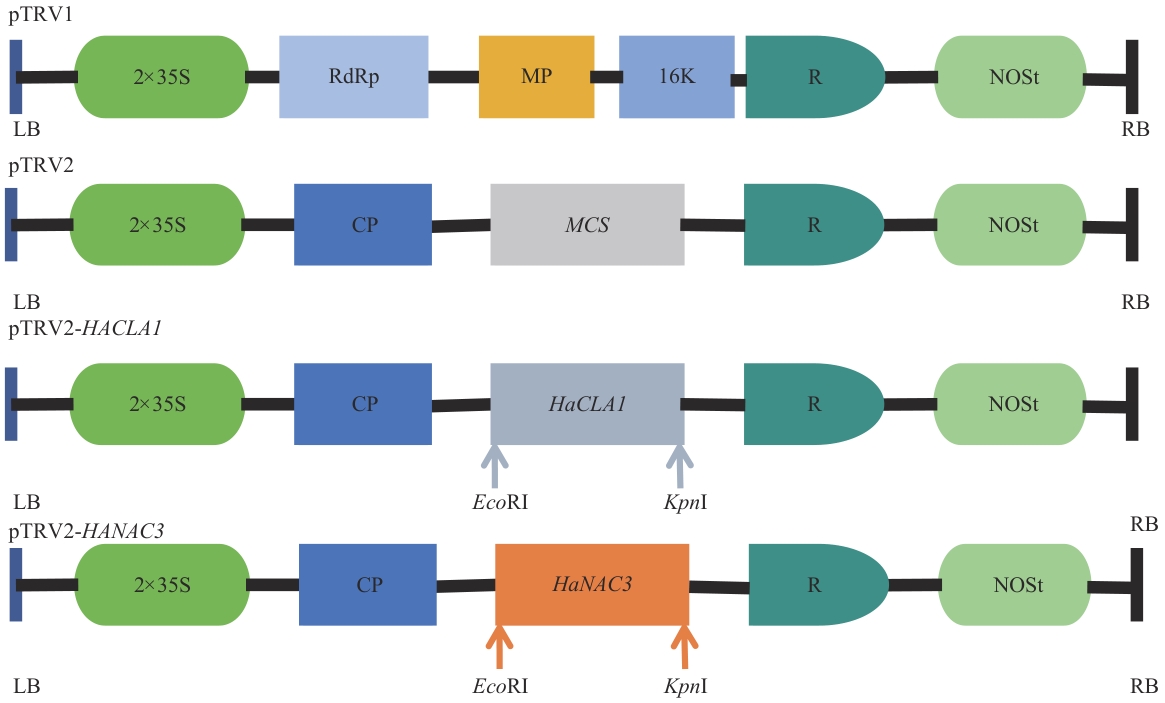
Fig. 1 Schematic diagram of pTRV1, pTRV2, pTRV2-HaCLA1and pTRV2-HaNAC3 vectorsLB: Left border. RB: Right border. 35S: CaMV 35S promoter. R: Self-cleaving ribozyme. NOSt: NOS terminator. CP: Viral capsid protein
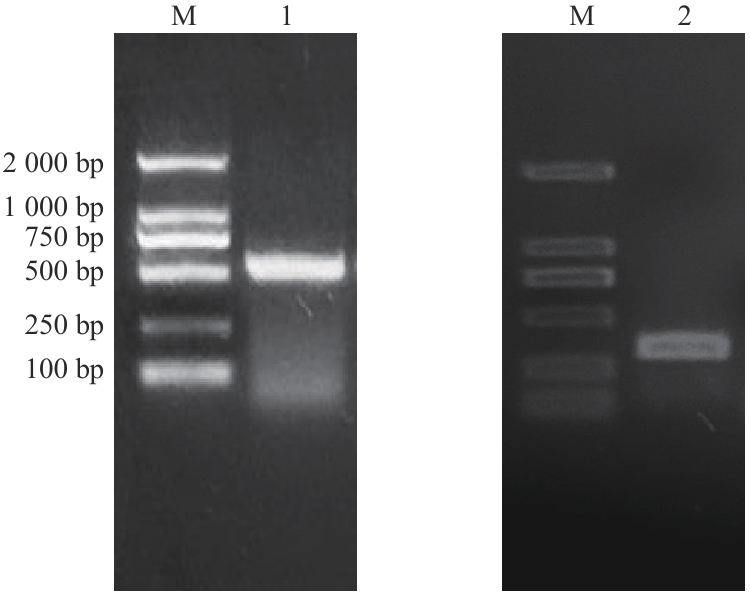
Fig. 3 Agarose gel electrophoresis verification of gene silencing fragment cloneM: 2 000 DNA marker. 1: HaCLA1 gene silencing fragment; 2: HaNAC3 gene silencing fragment

Fig. 4 Recombinant viral vector double enzyme digestion and verificationM:10 000 DNA marker;1:double digestion of pTRV2-HaCLA1 vector;2:pTRV2-HaCLA1 original plasmid electrophoresis
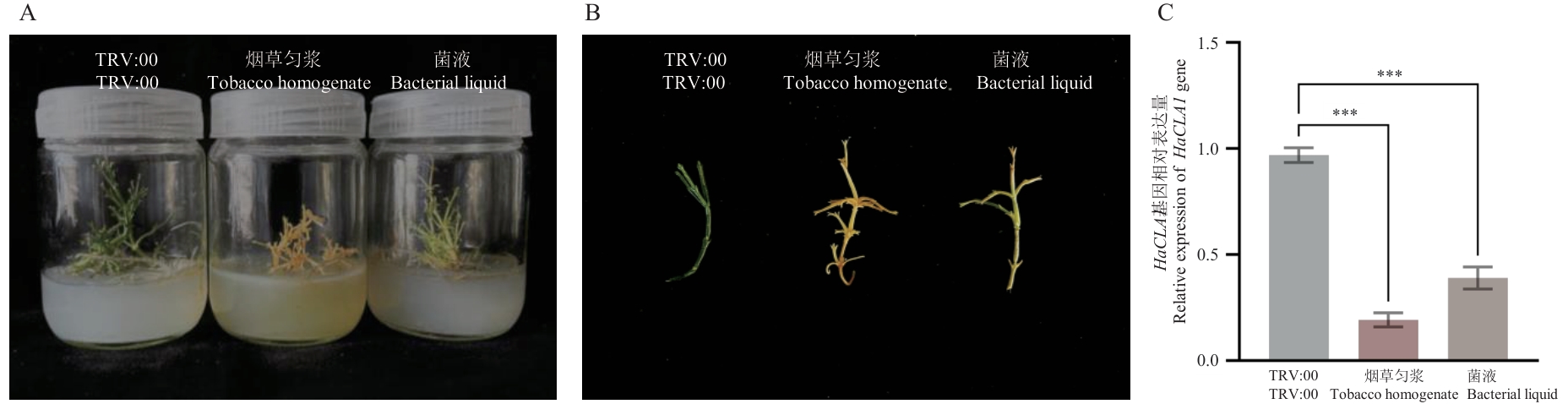
Fig. 5 Phenotype of HaCLA1 gene silencing and RT-qPCR analysis under different infection solutions (12 d of infestation)A:Whole-plant silencing phenotype of H. ammodendron after HaCLA1 gene silencing. B:Assimilating branch silencing phenotype of H. ammodendron after HaCLA1 gene silencing. C: Results of RT-qPCR analysis of HaCLA1 gene (12 d of infestation). ***P<0.001
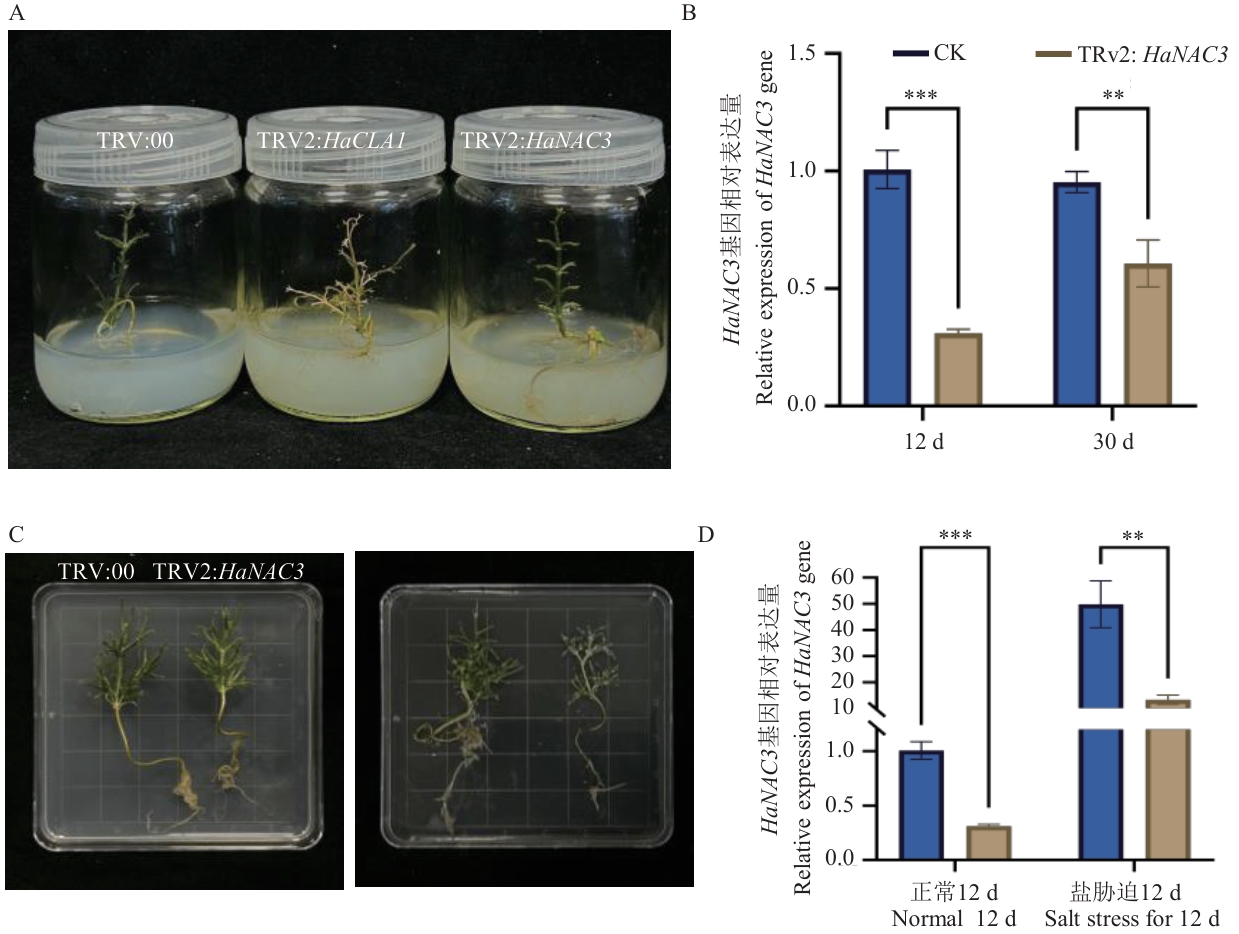
Fig. 6 Application of the H. ammodendron VIGS systemA: Phenotypic changes of HaNAC3 gene silencing in H. ammodendron. (TRV:00:Empty vector negative control group;TRV2:HaCLA1: Positive control group;TRV2:HaNAC3: Experimental group). B: Relative expression of the HaNAC3 gene in H. ammodendron after 12 d and 30 d. C: Phenotypic characteristics of H. ammodendron plants under normal conditions and after 12 d of salt stress. D: Relative expressions of the HaNAC3 gene in H. ammodendron under normal conditions and after 12 d of salt stress. ** indicates significant differences at the P<0.01, and *** indicates significant differences at the P<0.001
| [1] | Song J, Feng G, Tian CY, et al. Osmotic adjustment traits of Suaeda physophora, Haloxylon ammodendron and Haloxylon persicum in field or controlled conditions [J]. Plant Sci, 2006, 170(1): 113-119. |
| [2] | 张丹, 马松梅, 魏博, 等. 中国梭梭属植物历史分布格局及其驱动机制 [J]. 生物多样性, 2022, 30(1):42-51 |
| Zhang D, Ma SM, Wei B, et al. Historical distribution pattern and driving mechanism of Haloxylon in China [J]. Biodivers Sci, 2022, 30(1):42-51 | |
| [3] | 郑旭, 张倪斌, 孙桂丽, 等. 吐鲁番市高昌区不同固沙植物的防风固沙效益 [J]. 东北林业大学学报, 2024, 52(7): 44-50. |
| Zheng X, Zhang NB, Sun GL, et al. Wind prevention and sand-fixation benefits of different sand-fixing plants in Gaochang district, Turpan City [J]. J Northeast For Univ, 2024, 52(7): 44-50. | |
| [4] | 沈亮, 徐荣, 刘赛, 等. 肉苁蓉寄主梭梭根际土壤微生物种类及群落结构特征 [J]. 生态学报, 2016, 36(13): 3933-3942. |
| Shen L, Xu R, Liu S, et al. Characteristics of microbial community structure in rhizosphere soil of Haloxylon ammodendron [J]. Acta Ecol Sin, 2016, 36(13): 3933-3942. | |
| [5] | 许丽, 刘永刚, 王菊莲, 等. 石羊河下游荒漠区雨养梭梭柠条人工林健康评价 [J]. 防护林科技, 2025(1): 70-77. |
| Xu L, Liu YG, Wang JL, et al. Health evaluation of rain-fed Haloxylon ammodendron and Caragana korshinskii plantation in desert area of lower reaches of Shiyang River [J]. Prot For Sci Technol, 2025(1): 70-77. | |
| [6] | Liang JS, Liu XS, Xu L, et al. A novel NAC transcription factor from Haloxylon ammodendron promotes reproductive growth in Arabidopsis thaliana under drought stress [J]. Environ Exp Bot, 2024, 228: 106043. |
| [7] | Senthil-Kumar M, Mysore KS. New dimensions for VIGS in plant functional genomics [J]. Trends Plant Sci, 2011, 16(12): 656-665. |
| [8] | 潘多, 张嵩玥, 刘芳伊, 等. 病毒诱导的基因沉默技术用于植物色素代谢机制的研究进展 [J]. 生物工程学报, 2023, 39(7): 2579-2599. |
| Pan D, Zhang SY, Liu FY, et al. Application of virus-induced gene silencing technology to investigate the phytochrome metabolism mechanism: a review [J]. Chin J Biotechnol, 2023, 39(7): 2579-2599. | |
| [9] | Rosa C, Kuo YW, Wuriyanghan H, et al. RNA interference mechanisms and applications in plant pathology [J]. Annu Rev Phytopathol, 2018, 56: 581-610. |
| [10] | Yang W, Chen XY, Chen JH, et al. Virus-induced gene silencing in the tea plant (Camellia sinensis) [J]. Plants, 2023, 12(17): 3162. |
| [11] | 李瑞雪, 王钰婷, 胡飞, 等. 桑树PDS基因VIGS转化体系的构建与鉴定 [J]. 南方农业学报, 2018, 49(7): 1432-1438. |
| Li RX, Wang YT, Hu F, et al. VIGS transformation system construction and identification of gene PDS in mulberry [J]. J South Agric, 2018, 49(7): 1432-1438. | |
| [12] | Sasaki S, Yamagishi N, Yoshikawa N. Efficient virus-induced gene silencing in apple, pear and Japanese pear using Apple latent spherical virus vectors [J]. Plant Methods, 2011, 7(1): 15. |
| [13] | Cheuk A, Houde M. A rapid and efficient method for uniform gene expression using the barley stripe mosaic virus [J]. Plant Methods, 2017, 13: 24. |
| [14] | Mandel MA, Feldmann KA, Herrera-Estrella L, et al. CLA1 a novel gene required for chloroplast development, is highly conserved in evolution [J]. Plant J, 1996, 9(5): 649-658. |
| [15] | Senthil-Kumar M, Mysore KS. Tobacco rattle virus-based virus-induced gene silencing in Nicotiana benthamiana [J]. Nat Protoc, 2014, 9(7): 1549-1562. |
| [16] | Wang P, Man LJ, Ma L, et al. In vitro regeneration of Haloxylon ammodendron [J]. Not Sci Biol, 2023, 15(2): 11585. |
| [17] | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method [J]. Methods, 2001, 25(4): 402-408. |
| [18] | Chen PJ, Pang G, Cai F, et al. Strain improvement and genetic engineering of Trichoderma for industrial applications [M]//Encyclopedia of Mycology. Amsterdam: Elsevier, 2021: 505-517. |
| [19] | Azeez A, Bates PD. Self-incompatibility based functional genomics for rapid phenotypic characterization of seed metabolism genes [J]. Plant Biotechnol J, 2024, 22(10): 2688-2690. |
| [20] | Zhu YY, Yu XP, Wu J. CRISPR/Cas: a toolkit for plant disease diagnostics [J]. Trends Plant Sci, 2025, 30(3): 245-248. |
| [21] | Liu XS, Zong XF, Wu X, et al. Ectopic expression of NAC transcription factor HaNAC3 from Haloxylon ammodendron increased abiotic stress resistance in tobacco [J]. Planta, 2022, 256(6): 105. |
| [22] | Wang MC, Zhang L, Tong SF, et al. Chromosome-level genome assembly of a xerophytic plant, Haloxylon ammodendron [J]. DNA Res, 2022, 29(2): dsac006. |
| [23] | 张桦, 娜菲莎·木则帕尔, 任燕萍, 等.农杆菌介导的梭梭种子遗传转化体系构建的方法: CN118620955A [P]. 2024-09-10. |
| Zhang H, Nafeisha M, Ren YP, et al. Method for constructing a genetic transformation system of Haloxylon ammodendron seeds mediated by Agrobacterium: CN118620955A [P]. 2024-09-10. | |
| [24] | Cao YH, Ren W, Gao HJ, et al. HaASR2 from Haloxylon ammodendron confers drought and salt tolerance in plants [J]. Plant Sci, 2023, 328: 111572. |
| [25] | Navarro JA, Sanchez-Navarro JA, PALLAS V. Chapter One - Key checkpoints in the movement of plant viruses through the host [M] Advances in Virus Research: Vol. 104. Academic Press, 2019: 1-64. |
| [26] | Shi GY, Hao MY, Tian BM, et al. A newly established virus-induced gene silencing method via seed imbibition for functional genomics at early germination stages in cotton [J]. Ind Crops Prod, 2021, 172: 114040. |
| [27] | Scofield SR, Huang L, Brandt AS, et al. Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway [J]. Plant Physiol, 2005, 138(4): 2165-2173. |
| [28] | Kant R, Dasgupta I. Gene silencing approaches through virus-based vectors: speeding up functional genomics in monocots [J]. Plant Mol Biol, 2019, 100(1/2): 3-18. |
| [29] | Lu Y, Zeng FJ, Zhang ZH, et al. Differences in growth, ionomic and antioxidative enzymes system responded to neutral and alkali salt exposure in halophyte Haloxylon ammodendron seedlings [J]. Plant Physiol Biochem, 2025, 220: 109492. |
| [30] | Lü XP, Lü ZL, Zhang YM, et al. Lignin synthesis plays an essential role in the adaptation of Haloxylon ammodendron to adverse environments [J]. Int J Biol Macromol, 2025, 308: 142321. |
| [31] | 杨芳. 梭梭和白梭梭响应干旱的生理代谢和分子机制[D]. 乌鲁木齐: 新疆大学, 2022. |
| Yang F. Physiological metabolism and molecular mechanisms of Haloxylon ammodendron and Haloxylonpersicum in response to drought [D]. Urumqi: Xinjiang University, 2022 |
| [1] | HUANG Wen-jing, REN Si-chao, LIN Li, WANG You-ping, WU Jian. Advances in RNA Interference Technology for Plant Functional Genomics and Crop Improvement [J]. Biotechnology Bulletin, 2025, 41(9): 1-21. |
| [2] | LAI Shi-yu, LIANG Qiao-lan, WEI Lie-xin, NIU Er-bo, CHEN Ying-e, ZHOU Xin, YANG Si-zheng, WANG Bo. The Role of NbJAZ3 in the Infection of Nicotiana benthamiana by Alfalfa Mosaic Virus [J]. Biotechnology Bulletin, 2025, 41(8): 186-196. |
| [3] | NIU Jing-ping, ZHAO Jing, GUO Qian, WANG Shu-hong, ZHAO Jin-zhong, DU Wei-jun, YIN Cong-cong, YUE Ai-qin. Identification and Induced Expression Analysis of Transcription Factors NAC in Soybean Resistance to Soybean Mosaic Virus Based on WGCNA [J]. Biotechnology Bulletin, 2025, 41(7): 95-105. |
| [4] | LI Yi-jun, YANG Xiao-bei, XIA Lin, LUO Zhao-peng, XU Xin, YANG Jun, NING Qian-ji, WU Ming-zhu. Cloning and Functional Analysis of NtPRR37 Gene in Nicotiana tabacum L. [J]. Biotechnology Bulletin, 2024, 40(8): 221-231. |
| [5] | HUA Xuan, TIAN Bo-wen, ZHOU Xin-tong, JIANG Zi-han, WANG Shi-qi, HUANG Qian-hui, ZHANG Jian, CHEN Yan-hong. Cloning SmERF B3-45 from Salix matsudana and Functional Analysis on Its Tolerance to Salt [J]. Biotechnology Bulletin, 2024, 40(12): 124-135. |
| [6] | ZHANG Yi, ZHANG Xin-ru, ZHANG Jin-ke, HU Li-zong, SHANGGUAN Xin-xin, ZHENG Xiao-hong, HU Juan-juan, ZHANG Cong-cong, MU Gui-qing, LI Cheng-wei. Functional Analysis of TaMYB1 Gene in Wheat Under Cadmium Stress [J]. Biotechnology Bulletin, 2024, 40(1): 194-206. |
| [7] | LIU Zhen-yin, DUAN Zhi-zhen, PENG Ting, WANG Tong-xin, WANG Jian. Establishment and Optimization of Virus-induced Gene Silencing System in Bougainvillea peruviana ‘Thimma’ [J]. Biotechnology Bulletin, 2023, 39(7): 123-130. |
| [8] | FENG Ce-ting, JIANG Lyu, LIU Xin-ying, LUO Le, PAN Hui-tang, ZHANG Qi-xiang, YU Chao. Identification of the NAC Gene Family in Rosa persica and Response Analysis Under Drought Stress [J]. Biotechnology Bulletin, 2023, 39(11): 283-296. |
| [9] | ZHANG Yu-juan, LI Dong-hua, GONG Hui-hui, CUI Xin-xiao, GAO Chun-hua, ZHANG Xiu-rong, YOU Jun, ZHAO Jun-sheng. Cloning and Salt-tolerance Analysis of NAC Transcription Factor SiNAC77 from Sesamum indicum L. [J]. Biotechnology Bulletin, 2023, 39(11): 308-317. |
| [10] | LI Xiu-qing, HU Zi-yao, LEI Jian-feng, DAI Pei-hong, LIU Chao, DENG Jia-hui, LIU Min, SUN Ling, LIU Xiao-dong, LI Yue. Cloning and Functional Analysis of Gene GhTIFY9 Related to Cotton Verticillium Wilt Resistance [J]. Biotechnology Bulletin, 2022, 38(8): 127-134. |
| [11] | GAO Peng-fei, XI Fei-hu, ZHANG Ze-yu, HU Kai-qiang, CHEN Kai, WEI Wen-tao, DING Jia-zhi, GU Lian-feng. Research Progress of Plant VIGS Technology and Its Application in Forestry Science [J]. Biotechnology Bulletin, 2021, 37(5): 141-153. |
| [12] | WANG Fang, SUN Li-jiao, ZHAO Xiao-yu, WANG Jie-wan, SONG Xing-shun. Research Progresses on Plant NAC Transcription Factors [J]. Biotechnology Bulletin, 2019, 35(4): 88-93. |
| [13] | LEI Zhao-xia, LIU Jing, BAI Yi-ping, TANG Wei, WANG Hong-yang. Cloning and Functional Analysis of a Potato Ubiquitin-conjugating Enzyme Gene StUBC17 [J]. Biotechnology Bulletin, 2019, 35(1): 35-41. |
| [14] | YUAN Yi-hang, ZHANG He-hua, YOU Han-li, ZHANG Ling-yun. Cloning and Expression Analysis of PwNAC42 in Picea wilsonii [J]. Biotechnology Bulletin, 2018, 34(3): 113-120. |
| [15] | WANG Chun-yu, ZHANG Qian. Research Progress on Plant NAC Transcription Factors [J]. Biotechnology Bulletin, 2018, 34(11): 8-14. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||