








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (9): 1-21.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0645
HUANG Wen-jing( ), REN Si-chao, LIN Li, WANG You-ping, WU Jian(
), REN Si-chao, LIN Li, WANG You-ping, WU Jian( )
)
Received:2025-06-19
Online:2025-09-26
Published:2025-09-24
Contact:
WU Jian
E-mail:222101206@stu.yzu.edu.cn;wu_jian@yzu.edu.cn
HUANG Wen-jing, REN Si-chao, LIN Li, WANG You-ping, WU Jian. Advances in RNA Interference Technology for Plant Functional Genomics and Crop Improvement[J]. Biotechnology Bulletin, 2025, 41(9): 1-21.

Fig. 1 Discovery history of RNA interferenceFrom left to right, Figure 1 presents key milestones in the discovery of RNA interference technology and the formation of its theoretical basis from 1989 to 2006: ranging from the discovery that nucleic acid sequence homology regulates gene expression, to clarification of dsRNA-mediated gene silencing, and finally to the recognition of its significance by the Nobel Prize. All figures were created in https://Biorender.com. The same below
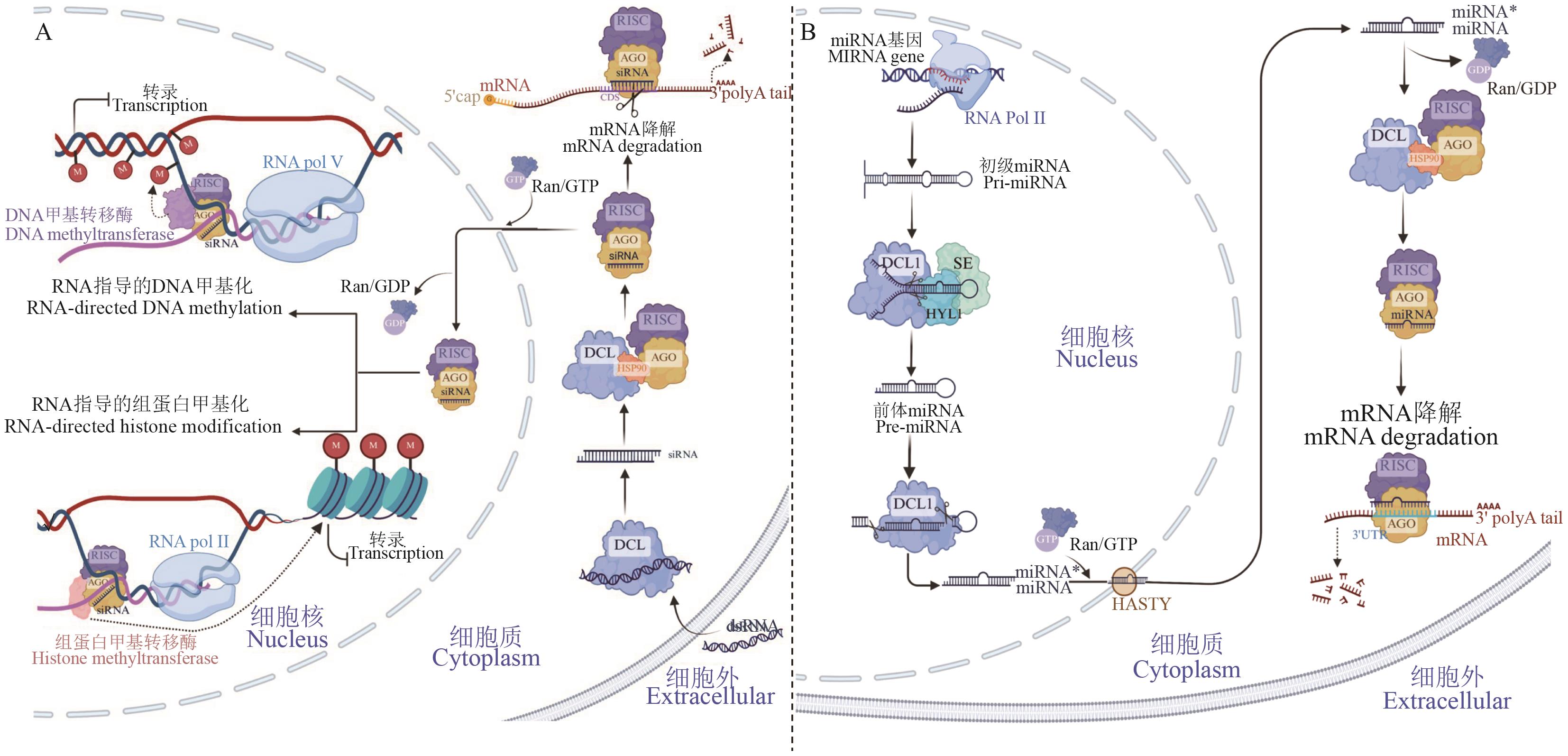
Fig. 2 Mechanism of RNA interferenceA: Mechanism of RNA interference (RNAi) mediated by exogenous small interfering RNA (siRNA). Dicer-like (DCL) cleaves exogenous dsRNA into siRNAs, which are then assembled into Argonaute (AGO)-containing RNA-induced silencing complex (RISC). In the cytoplasm, RISC binds to target mRNA through siRNA complementarity and cleaves it (post-transcriptional silencing). In the nucleus, siRNA-AGO complexes bind to lncRNAs transcribed by RNA polymerase V, mediating RNA-directed DNA or histone methylation (transcriptional silencing). B: Mechanism of RNAi mediated by endogenous microRNA (miRNA) in the nucleus, the DCL1 complex processes pri-miRNA into pre-miRNA and miRNA duplexes, which are then transported to the cytoplasm via HASTY. The mature miRNA is loaded into RISC, and AGO mediates the cleavage of target mRNA (with cleavage being the primary mechanism in plants)

Fig. 3 Mechanisms of virus-induced gene silencing (VIGS) mediated by different virusesThe left figure illustrates the mechanism of virus-induced gene silencing (VIGS) mediated by single-stranded DNA (ssDNA) viruses: Following receptor-mediated entry into the cell and uncoating, the viral ssDNA enters the nucleus, where it is converted into circular double-stranded DNA (dsDNA) through rolling-circle replication using host enzymes. This dsDNA is subsequently processed by DCL3 into 24-nucleotide (nt) siRNAs, which guide the RISC-AGO4 complex to mediate DNA methylation and thereby suppress transcription. The right figure depicts the VIGS mechanism mediated by positive-sense single-stranded RNA (+ssRNA) viruses: After entering the cell, the viral +ssRNA functions as mRNA to translate replicases. During replication, the formed dsRNA and local double-stranded regions are processed by DCL4/DCL2 into 21/22-nt siRNAs, which then mediate mRNA degradation and translation inhibition via RISC

Fig. 4 Mechanisms of RNAi in host-induced gene silencing (HIGS) and spray-induced gene silencing (SIGS)A: The mechanism of host-induced gene silencing (HIGS) is presented from top to bottom as follows: When chewing insects feed on plants, they ingest long dsRNAs in chloroplasts that have not been cleaved by DCL. These dsRNAs enter cells through endocytosis, SID protein-mediated pathways, etc., triggering RNAi to silence target genes. When piercing-sucking insects feed on phloem (containing siRNAs and dsRNAs), RNAi is triggered to silence target genes. During fungal/viral infection, plants deliver sRNAs through vesicles, or process hpRNAs into dsRNAs/siRNAs themselves, thereby trans-kingdom silencing pathogenic genes (the details of delivery remain to be clarified). B: Two pathways of spray-induced gene silencing (SIGS) technology for disease control: Firstly, pathogenic cells directly absorb naked dsRNAs in the environment; secondly, plant-mediated transport: after being absorbed by plants, dsRNAs are transported over long distances through the vascular system, processed into siRNAs or retained as dsRNAs by DCL, and then delivered to pathogens via extracellular vesicles (EVs) to trigger gene silencing. Question marks indicate unelucidated small RNA uptake mechanisms
病原体 Pathogen | 寄主植物 Host plant | 靶标基因 Target gene | 结果 Results | 参考文献 Reference |
|---|---|---|---|---|
大麦白粉病菌 Puccinia spp. | 大麦 | 1,3-β-葡聚糖转移酶(GTF1/2)效应子Avral0 | 白粉病菌吸器减少并抑制菌丝生长,大麦抗性提高 | [ |
| 大麦 | 效应子(BEC101、C1054等8个基因) | 干扰真菌分泌蛋白,阻断致病信号传导,抑制孢子萌发和侵染 | [ | |
小麦叶锈病菌 P. triticina | 小麦 | 促分裂素原活化蛋白激酶(PtMAPK1)、亲环蛋白(PtCYC1)、钙依赖磷酸酶B基因(PtCNB) | 病原菌中靶基因转录水平降低59%-70%,菌丝生长受限、吸器发育异常,夏孢子堆密度减少51%-68% | [ |
| 小麦 | 丝裂原活化蛋白激酶1基因(PtMAPK1)、亲环蛋白基因(PtCYC1) | 干扰真菌细胞周期和应激反应,限制菌丝扩展,真菌生物量减少超50% | [ | |
小麦条锈病菌 P. striiformis | 小麦 | 蛋白激酶A催化亚基基因(PsCPK1) | 阻断cAMP信号通路,抑制菌丝极性生长和吸器形成,稳定抗病性持续4代 | [ |
| 小麦 | 丝裂原活化蛋白激酶激酶基因(PsFuz7) | 抑制MAPK信号传导,显著限制病原菌菌丝发育 | [ | |
禾谷镰刀菌 Fusarium graminearum | 大麦 | 麦角固醇合成相关的甾醇-14a-去甲基化酶基因(CYP51-A、CYP51-B和CYP51-C) | 靶基因表达抑制率77%-92%,真菌菌丝生长被限制在接种部位 | [ |
| 小麦 | 丁质合成酶3b基因(Chs3b) | 阻断细胞壁几丁质合成,抑制分生孢子萌发和侵染,降低DON毒素产生 | [ | |
核盘菌 Sclerotinia sclerotiorum | 油菜 | 内切多聚半乳糖醛酸酶基因(SsPG1)、纤维二糖水解酶基因(SsCBH)和草酰乙酸乙酰水解酶基因(SsOAH1) | 单靶标HIGS:病斑面积减少20.8%-38.7% 三靶标共沉默HIGS:病斑面积减少36.8%-43.7%,抗性显著增强 | [ |
| 油菜 | 内切多聚半乳糖醛酸酶基因(SsPG1) | 子叶病斑面积减少51.8%-58.2%,叶片病斑面积减少20.1%-26.2% | [ | |
灰霉菌 Botrytis cinerea | 番茄 | 灰霉病菌致病基因Bc-DCL1和Bc-DCL2 | 靶基因表达量下降70%-90%,病斑面积减少50%-80%,孢子萌发率下降30%-60% | [ |
马铃薯 番茄 | 雷帕霉素靶蛋白基因(BcTOR) | 靶基因表达下调60%-90%,病斑面积减少50%- 90%,真菌生物量降低60%-85%,抑制灰霉病发展 | [ | |
辣椒疫霉 Phytophthora capsici | 烟草 | 纤维素合成酶基因(PcCesA3)、固醇结合蛋白基因(PcOSBP1) | 病斑面积减少17.19%-33.34%,病原菌生物量降低59.64%-77.65% | [ |
立枯丝核菌 Rhizoctonia solani | 水稻 | 效应子基因AGLIP1 | 抑制菌丝生长和致病力,使接种后病斑面积减少50% | [ |
葡萄座腔菌 Botryosphaeria dothidea | 湖北海棠 | 真菌糖转运蛋白基因(BdSTP)、乙酰乳酸合成酶基因(BdALS) | 病斑面积减少超50%,真菌生物量降低约60%-70% | [ |
棉铃虫 Helicoverpa armigera | 烟草 | 棉蜕皮激素受体基因(EcR) | EcR基因表达显著下调,出现蜕皮缺陷和致死表型;对甜菜夜蛾也有抗性 | [ |
| 二斑叶螨Tetranychus urticae | 烟草 | 保幼激素受体基因Met | 二斑叶螨死亡率达48% | [ |
| 桃蚜Myzus persicae | 烟草 | V-ATPase亚基E、微管蛋白折叠辅助因子D(TBCD)或乙酰胆碱酯酶2(MpAchE2) | 单只成虫产生的若虫数量减少约30% | [ |
| 短体线虫Pratylenchus | 小麦 | 肌钙蛋白C(pat-10)、钙调蛋白(unc-87) | 线虫瘫痪和不协调运动,减少繁殖 | [ |
| 南方根结线虫Meloidogyne incognita | 番茄 | 组织蛋白酶L半胱氨酸蛋白酶(Mi-cpl-1) | 线虫感染和增殖减少60%-80% | [ |
| 西方玉米根虫Diabrotica virgifera virgifera | 玉米 | V-ATPase亚基A或C | 减少对根系的损害 | [ |
| 粉虱Aleyrodidae | 烟草 | 粉虱V-ATPaseA基因 | 植物定殖减少,粉虱死亡率增加 | [ |
| 麦长管蚜Sitobion avenae | 小麦 | 羧酸酯酶基因(CbEE4) | 基因表达量降低30%-60%,羧酸酯酶活性下降, 对辛硫磷农药水解能力降至20%-30%,蚜虫繁殖减少 | [ |
| 香蕉束顶病毒Banana bunchy top virus | 香蕉 | 复制酶基因(rep) | 转基因植株对BBTV完全免疫、病毒复制几乎完全被抑制 | [ |
| 玉米矮花叶病毒Maize dwarf mosaic virus | 玉米 | 蛋白酶基因(P1) | 15个T₂代株系均增强抗病性,其中6个株系病斑指数低于25%,病毒P1基因相对复制水平显著降低 | [ |
| 水稻黑条矮缩病毒Rice black-streaked dwarf virus | 水稻 | 水稻黑条矮缩病毒的P7-2和P8基因 | 转基因后代表现出较强抗病毒性 | [ |
| 小麦条纹花叶病毒Wheat streak mosaic virus | 小麦 | 小麦条纹花叶病毒外壳蛋白基因 | 通过RNAi抑制病毒衣壳组装、阻止病毒颗粒形成,持续抗性至T5代 | [ |
| 烟草条纹病毒Tobacco streak virus | 烟草 | 烟草条纹病毒复制酶部分序列 | 诱导病毒基因沉默,使其获得抗TSV特性 | [ |
| 柑橘衰退病毒Citrus tristeza virus | 柑橘 | 沉默抑制蛋白基因p25、p20和p23 | 3个转基因株系对CTV-T36完全抗病,嫁接接种后无症状且无病毒 | [ |
| 李痘病毒Plum pox virus | 欧洲李、烟草 | 外壳蛋白基因(CP) | 对PPV主要株系(D、M、Rec、EA)表现出完全抗性,病毒未发生系统性扩散 | [ |
Table 1 Advances in research on HIGS technology in disease control and prevention
病原体 Pathogen | 寄主植物 Host plant | 靶标基因 Target gene | 结果 Results | 参考文献 Reference |
|---|---|---|---|---|
大麦白粉病菌 Puccinia spp. | 大麦 | 1,3-β-葡聚糖转移酶(GTF1/2)效应子Avral0 | 白粉病菌吸器减少并抑制菌丝生长,大麦抗性提高 | [ |
| 大麦 | 效应子(BEC101、C1054等8个基因) | 干扰真菌分泌蛋白,阻断致病信号传导,抑制孢子萌发和侵染 | [ | |
小麦叶锈病菌 P. triticina | 小麦 | 促分裂素原活化蛋白激酶(PtMAPK1)、亲环蛋白(PtCYC1)、钙依赖磷酸酶B基因(PtCNB) | 病原菌中靶基因转录水平降低59%-70%,菌丝生长受限、吸器发育异常,夏孢子堆密度减少51%-68% | [ |
| 小麦 | 丝裂原活化蛋白激酶1基因(PtMAPK1)、亲环蛋白基因(PtCYC1) | 干扰真菌细胞周期和应激反应,限制菌丝扩展,真菌生物量减少超50% | [ | |
小麦条锈病菌 P. striiformis | 小麦 | 蛋白激酶A催化亚基基因(PsCPK1) | 阻断cAMP信号通路,抑制菌丝极性生长和吸器形成,稳定抗病性持续4代 | [ |
| 小麦 | 丝裂原活化蛋白激酶激酶基因(PsFuz7) | 抑制MAPK信号传导,显著限制病原菌菌丝发育 | [ | |
禾谷镰刀菌 Fusarium graminearum | 大麦 | 麦角固醇合成相关的甾醇-14a-去甲基化酶基因(CYP51-A、CYP51-B和CYP51-C) | 靶基因表达抑制率77%-92%,真菌菌丝生长被限制在接种部位 | [ |
| 小麦 | 丁质合成酶3b基因(Chs3b) | 阻断细胞壁几丁质合成,抑制分生孢子萌发和侵染,降低DON毒素产生 | [ | |
核盘菌 Sclerotinia sclerotiorum | 油菜 | 内切多聚半乳糖醛酸酶基因(SsPG1)、纤维二糖水解酶基因(SsCBH)和草酰乙酸乙酰水解酶基因(SsOAH1) | 单靶标HIGS:病斑面积减少20.8%-38.7% 三靶标共沉默HIGS:病斑面积减少36.8%-43.7%,抗性显著增强 | [ |
| 油菜 | 内切多聚半乳糖醛酸酶基因(SsPG1) | 子叶病斑面积减少51.8%-58.2%,叶片病斑面积减少20.1%-26.2% | [ | |
灰霉菌 Botrytis cinerea | 番茄 | 灰霉病菌致病基因Bc-DCL1和Bc-DCL2 | 靶基因表达量下降70%-90%,病斑面积减少50%-80%,孢子萌发率下降30%-60% | [ |
马铃薯 番茄 | 雷帕霉素靶蛋白基因(BcTOR) | 靶基因表达下调60%-90%,病斑面积减少50%- 90%,真菌生物量降低60%-85%,抑制灰霉病发展 | [ | |
辣椒疫霉 Phytophthora capsici | 烟草 | 纤维素合成酶基因(PcCesA3)、固醇结合蛋白基因(PcOSBP1) | 病斑面积减少17.19%-33.34%,病原菌生物量降低59.64%-77.65% | [ |
立枯丝核菌 Rhizoctonia solani | 水稻 | 效应子基因AGLIP1 | 抑制菌丝生长和致病力,使接种后病斑面积减少50% | [ |
葡萄座腔菌 Botryosphaeria dothidea | 湖北海棠 | 真菌糖转运蛋白基因(BdSTP)、乙酰乳酸合成酶基因(BdALS) | 病斑面积减少超50%,真菌生物量降低约60%-70% | [ |
棉铃虫 Helicoverpa armigera | 烟草 | 棉蜕皮激素受体基因(EcR) | EcR基因表达显著下调,出现蜕皮缺陷和致死表型;对甜菜夜蛾也有抗性 | [ |
| 二斑叶螨Tetranychus urticae | 烟草 | 保幼激素受体基因Met | 二斑叶螨死亡率达48% | [ |
| 桃蚜Myzus persicae | 烟草 | V-ATPase亚基E、微管蛋白折叠辅助因子D(TBCD)或乙酰胆碱酯酶2(MpAchE2) | 单只成虫产生的若虫数量减少约30% | [ |
| 短体线虫Pratylenchus | 小麦 | 肌钙蛋白C(pat-10)、钙调蛋白(unc-87) | 线虫瘫痪和不协调运动,减少繁殖 | [ |
| 南方根结线虫Meloidogyne incognita | 番茄 | 组织蛋白酶L半胱氨酸蛋白酶(Mi-cpl-1) | 线虫感染和增殖减少60%-80% | [ |
| 西方玉米根虫Diabrotica virgifera virgifera | 玉米 | V-ATPase亚基A或C | 减少对根系的损害 | [ |
| 粉虱Aleyrodidae | 烟草 | 粉虱V-ATPaseA基因 | 植物定殖减少,粉虱死亡率增加 | [ |
| 麦长管蚜Sitobion avenae | 小麦 | 羧酸酯酶基因(CbEE4) | 基因表达量降低30%-60%,羧酸酯酶活性下降, 对辛硫磷农药水解能力降至20%-30%,蚜虫繁殖减少 | [ |
| 香蕉束顶病毒Banana bunchy top virus | 香蕉 | 复制酶基因(rep) | 转基因植株对BBTV完全免疫、病毒复制几乎完全被抑制 | [ |
| 玉米矮花叶病毒Maize dwarf mosaic virus | 玉米 | 蛋白酶基因(P1) | 15个T₂代株系均增强抗病性,其中6个株系病斑指数低于25%,病毒P1基因相对复制水平显著降低 | [ |
| 水稻黑条矮缩病毒Rice black-streaked dwarf virus | 水稻 | 水稻黑条矮缩病毒的P7-2和P8基因 | 转基因后代表现出较强抗病毒性 | [ |
| 小麦条纹花叶病毒Wheat streak mosaic virus | 小麦 | 小麦条纹花叶病毒外壳蛋白基因 | 通过RNAi抑制病毒衣壳组装、阻止病毒颗粒形成,持续抗性至T5代 | [ |
| 烟草条纹病毒Tobacco streak virus | 烟草 | 烟草条纹病毒复制酶部分序列 | 诱导病毒基因沉默,使其获得抗TSV特性 | [ |
| 柑橘衰退病毒Citrus tristeza virus | 柑橘 | 沉默抑制蛋白基因p25、p20和p23 | 3个转基因株系对CTV-T36完全抗病,嫁接接种后无症状且无病毒 | [ |
| 李痘病毒Plum pox virus | 欧洲李、烟草 | 外壳蛋白基因(CP) | 对PPV主要株系(D、M、Rec、EA)表现出完全抗性,病毒未发生系统性扩散 | [ |
| [89] | Lomate PR, Bonning BC. Distinct properties of proteases and nucleases in the gut, salivary gland and saliva of southern green stink bug, Nezara viridula [J]. Sci Rep, 2016, 6: 27587. |
| [90] | Koch A, Wassenegger M. Host-induced gene silencing - mechanisms and applications [J]. New Phytol, 2021, 231(1): 54-59. |
| [91] | Qiao LL, Lan C, Capriotti L, et al. Spray-induced gene silencing for disease control is dependent on the efficiency of pathogen RNA uptake [J]. Plant Biotechnol J, 2021, 19(9): 1756-1768. |
| [92] | Wang M, Weiberg A, Lin FM, et al. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection [J]. Nat Plants, 2016, 2: 16151. |
| [93] | Koch A, Biedenkopf D, Furch A, et al. An RNAi-based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery [J]. PLoS Pathog, 2016, 12(10): e1005901. |
| [94] | Cai Q, Qiao LL, Wang M, et al. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes [J]. Science, 2018, 360(6393): 1126-1129. |
| [95] | Zhao DY, Zhong GY, Song GQ. Transfer of endogenous small RNAs between branches of scions and rootstocks in grafted sweet cherry trees [J]. PLoS One, 2020, 15(7): e0236376. |
| [96] | Pliego C, Nowara D, Bonciani G, et al. Host-induced gene silencing in barley powdery mildew reveals a class of ribonuclease-like effectors [J]. Mol Plant Microbe Interact, 2013, 26(6): 633-642. |
| [97] | Panwar V, McCallum B, Bakkeren G. Endogenous silencing of Puccinia triticina pathogenicity genes through in planta-expressed sequences leads to the suppression of rust diseases on wheat [J]. Plant J, 2013, 73(3): 521-532. |
| [98] | Panwar V, Jordan M, McCallum B, et al. Host-induced silencing of essential genes in Puccinia triticina through transgenic expression of RNAi sequences reduces severity of leaf rust infection in wheat [J]. Plant Biotechnol J, 2018, 16(5): 1013-1023. |
| [99] | Qi T, Zhu XG, Tan CL, et al. Host-induced gene silencing of an important pathogenicity factor PsCPK1 in Puccinia striiformis f. sp. tritici enhances resistance of wheat to stripe rust [J]. Plant Biotechnol J, 2018, 16(3): 797-807. |
| [100] | Zhu XG, Qi T, Yang Q, et al. Host-induced gene silencing of the MAPKK gene PsFUZ7 confers stable resistance to wheat stripe rust [J]. Plant Physiol, 2017, 175(4): 1853-1863. |
| [101] | Koch A, Kumar N, Weber L, et al. Host-induced gene silencing of cytochrome P450 lanosterol C14α-demethylase-encoding genes confers strong resistance to Fusarium species [J]. Proc Natl Acad Sci USA, 2013, 110(48): 19324-19329. |
| [102] | Cheng W, Song XS, Li HP, et al. Host-induced gene silencing of an essential chitin synthase gene confers durable resistance to Fusarium head blight and seedling blight in wheat [J]. Plant Biotechnol J, 2015, 13(9): 1335-1345. |
| [103] | Xiong FJ, Liu M, Zhuo FP, et al. Host-induced gene silencing of BcTOR in Botrytis cinerea enhances plant resistance to grey mould [J]. Mol Plant Pathol, 2019, 20(12): 1722-1739. |
| [104] | Wu J, Yin SL, Lin L, et al. Host-induced gene silencing of multiple pathogenic factors of Sclerotinia sclerotiorum confers resistance to Sclerotinia rot in Brassica napus [J]. Crop J, 2022, 10(3): 661-671. |
| [105] | Lin L, Fan JL, Li PP, et al. The Sclerotinia sclerotiorum-inducible promoter pBnGH17D7 in Brassica napus: isolation, characterization, and application in host-induced gene silencing [J]. J Exp Bot, 2022, 73(19): 6663-6677. |
| [106] | Wang ZW, Gao X, Zhong S, et al. Host-induced gene silencing of PcCesA3 and PcOSBP1 confers resistance to Phytophthora capsici in Nicotiana benthamiana through NbDCL3 and NbDCL4 processed small interfering RNAs [J]. Int J Biol Macromol, 2022, 222: 1665-1675. |
| [107] | Zhao M, Liu XX, Wan J, et al. Host-induced gene silencing of effector AGLIP1 enhanced resistance of rice to Rhizoctonia solani AG1-IA [J]. Rice Sci, 2024, 31(4): 463-474. |
| [108] | Yu XY, Lin XX, Zhou TT, et al. Host-induced gene silencing in wild apple germplasm Malus hupehensis confers resistance to the fungal pathogen Botryosphaeria dothidea [J]. Plant J, 2024, 118(4): 1174-1193. |
| [109] | Zhu JQ, Liu SM, Ma Y, et al. Improvement of pest resistance in transgenic tobacco plants expressing dsRNA of an insect-associated gene EcR [J]. PLoS One, 2012, 7(6): e38572. |
| [110] | Yoon JS, Sahoo DK, Maiti IB, et al. Identification of target genes for RNAi-mediated control of the twospotted spider mite [J]. Sci Rep, 2018, 8(1): 14687. |
| [111] | Xu LJ, Duan XL, Lv YH, et al. Silencing of an aphid carboxylesterase gene by use of plant-mediated RNAi impairs Sitobion avenae tolerance of Phoxim insecticides [J]. Transgenic Res, 2014, 23(2): 389-396. |
| [112] | Guo HY, Song XG, Wang GL, et al. Plant-generated artificial small RNAs mediated aphid resistance [J]. PLoS One, 2014, 9(5): e97410. |
| [113] | Tan JCH, Jones MGK, Fosu-Nyarko J. Gene silencing in root lesion nematodes (Pratylenchus spp.) significantly reduces reproduction in a plant host [J]. Exp Parasitol, 2013, 133(2): 166-178. |
| [114] | Dutta TK, Banakar P, Rao U. The status of RNAi-based transgenic research in plant nematology [J]. Front Microbiol, 2015, 5: 760. |
| [115] | Li H, Khajuria C, Rangasamy M, et al. Long dsRNA but not siRNA initiates RNAi in western corn rootworm larvae and adults [J]. J Appl Entomol, 2015, 139(6): 432-445. |
| [116] | Thakur N, Upadhyay SK, Verma PC, et al. Enhanced whitefly resistance in transgenic tobacco plants expressing double stranded RNA of v-ATPase A gene [J]. PLoS One, 2014, 9(3): e87235. |
| [117] | Elayabalan S, Kalaiponmani K, Subramaniam S, et al. Development of Agrobacterium-mediated transformation of highly valued hill banana cultivar Virupakshi (AAB) for resistance to BBTV disease [J]. World J Microbiol Biotechnol, 2013, 29(4): 589-596. |
| [118] | Zhang ZY, Wang YG, Shen XJ, et al. RNA interference-mediated resistance to maize dwarf mosaic virus [J]. Plant Cell Tissue Organ Cult, 2013, 113(3): 571-578. |
| [119] | Ahmed MMS, Bian SQ, Wang MY, et al. RNAi-mediated resistance to rice black-streaked dwarf virus in transgenic rice [J]. Transgenic Res, 2017, 26(2): 197-207. |
| [120] | Cruz LF, Rupp JLS, Trick HN, et al. Stable resistance to Wheat streak mosaic virus in wheat mediated by RNAi [J]. Vitro Cell Dev Biol Plant, 2014, 50(6): 665-672. |
| [121] | Suppaiah R, Muthuraj R, Gandhi K. Conserved sequence of replicase gene mediated resistance in Nicotiana tabacum L. cv Abirami through RNA silencing [J]. Eur J Plant Pathol, 2015, 142(4): 865-874. |
| [122] | Soler N, Plomer M, Fagoaga C, et al. Transformation of Mexican lime with an intron-hairpin construct expressing untranslatable versions of the genes coding for the three silencing suppressors of Citrus tristeza virus confers complete resistance to the virus [J]. Plant Biotechnol J, 2012, 10(5): 597-608. |
| [123] | Ravelonandro M, Scorza R, Michel HJ, et al. The efficiency of RNA interference for conferring stable resistance to plum pox virus [J]. Plant Cell Tissue Organ Cult, 2014, 118(2): 347-356. |
| [124] | Lück S, Kreszies T, Strickert M, et al. siRNA-Finder (si-Fi) software for RNAi-target design and off-target prediction [J]. Front Plant Sci, 2019, 10: 1023. |
| [1] | Matzke MA, Primig M, Trnovsky J, et al. Reversible methylation and inactivation of marker genes in sequentially transformed tobacco plants [J]. EMBO J, 1989, 8(3): 643-649. |
| [2] | Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric Chalcone synthase gene into Petunia results in reversible co-suppression of homologous genes in trans [J]. Plant Cell, 1990, 2(4): 279-289. |
| [3] | Fire A, Albertson D, Harrison SW, et al. Production of antisense RNA leads to effective and specific inhibition of gene expression in C. elegans muscle [J]. Development, 1991, 113(2): 503-514. |
| [4] | Romano N, Macino G. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences [J]. Mol Microbiol, 1992, 6(22): 3343-3353. |
| [5] | Guo S, Kemphues KJ. Par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed [J]. Cell, 1995, 81(4): 611-620. |
| [6] | Fire A, Xu S, Montgomery MK, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans [J]. Nature, 1998, 391(6669): 806-811. |
| [7] | Bernstein E, Caudy AA, Hammond SM, et al. Role for a bidentate ribonuclease in the initiation step of RNA interference [J]. Nature, 2001, 409(6818): 363-366. |
| [8] | Elbashir SM, Harborth J, Lendeckel W, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells [J]. Nature, 2001, 411(6836): 494-498. |
| [9] | Filipowicz W. RNAi: the nuts and bolts of the RISC machine [J]. Cell, 2005, 122(1): 17-20. |
| [10] | Iki T, Yoshikawa M, Nishikiori M, et al. In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90 [J]. Mol Cell, 2010, 39(2): 282-291. |
| [11] | Hammond SM, Bernstein E, Beach D, et al. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells [J]. Nature, 2000, 404(6775): 293-296. |
| [12] | Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase pol IVb/pol V mediates transcriptional silencing of overlapping and adjacent genes [J]. Cell, 2008, 135(4): 635-648. |
| [13] | Matzke MA, Mosher RA. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity [J]. Nat Rev Genet, 2014, 15(6): 394-408. |
| [14] | Weinberg MS, Villeneuve LM, Ehsani A, et al. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells [J]. RNA, 2006, 12(2): 256-262. |
| [15] | Grewal SIS, Elgin SCR. Transcription and RNA interference in the formation of heterochromatin [J]. Nature, 2007, 447(7143): 399-406. |
| [16] | Song L, Han MH, Lesicka J, et al. Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body [J]. Proc Natl Acad Sci USA, 2007, 104(13): 5437-5442. |
| [17] | Gregory RI, Yan KP, Amuthan G, et al. The Microprocessor complex mediates the genesis of microRNAs [J]. Nature, 2004, 432(7014): 235-240. |
| [18] | Bologna NG, Schapire AL, Zhai JX, et al. Multiple RNA recognition patterns during microRNA biogenesis in plants [J]. Genome Res, 2013, 23(10): 1675-1689. |
| [19] | Park MY, Wu G, Gonzalez-Sulser A, et al. Nuclear processing and export of microRNAs in Arabidopsis [J]. Proc Natl Acad Sci USA, 2005, 102(10): 3691-3696. |
| [20] | Schwarz DS, Hutvágner G, Du TT, et al. Asymmetry in the assembly of the RNAi enzyme complex [J]. Cell, 2003, 115(2): 199-208. |
| [21] | Llave C, Kasschau KD, Rector MA, et al. Endogenous and silencing-associated small RNAs in plants [J]. Plant Cell, 2002, 14(7): 1605-1619. |
| [22] | German MA, Pillay M, Jeong DH, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends [J]. Nat Biotechnol, 2008, 26(8): 941-946. |
| [23] | Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs [J]. Nature, 2005, 433(7027): 769-773. |
| [24] | Aregger M, Borah BK, Seguin J, et al. Primary and secondary siRNAs in geminivirus-induced gene silencing [J]. PLoS Pathog, 2012, 8(9): e1002941. |
| [125] | Xu Y, Tan JY, Lu JX, et al. RAS signalling genes can be used as host-induced gene silencing targets to control fungal diseases caused by Sclerotinia sclerotiorum and Botrytis cinerea [J]. Plant Biotechnol J, 2024, 22(1): 262-277. |
| [126] | Fahim M, Millar AA, Wood CC, et al. Resistance to Wheat streak mosaic virus generated by expression of an artificial polycistronic microRNA in wheat [J]. Plant Biotechnol J, 2012, 10(2): 150-163. |
| [127] | Xie HT, Gan P, Lü SY, et al. Development of marker-free transgenic rice exhibiting stable and enhanced resistance to Rice ragged stunt virus and Rice grassy stunt virus via RNA interference [J]. Plant Biotechnol J, 2024, 22(8): 2327-2329. |
| [128] | Wang PL, Si H, Li CH, et al. Plant genetic transformation: achievements, current status and future prospects [J]. Plant Biotechnol J, 2025, 23(6): 2034-2058. |
| [129] | Head GP, Carroll MW, Evans SP, et al. Evaluation of SmartStax and SmartStax PRO maize against western corn rootworm and northern corn rootworm: efficacy and resistance management [J]. Pest Manag Sci, 2017, 73(9): 1883-1899. |
| [130] | He L, Huang YN, Tang XM. RNAi-based pest control: production, application and the fate of dsRNA [J]. Front Bioeng Biotechnol, 2022, 10: 1080576. |
| [131] | Cappelle K, de Oliveira CFR, Van Eynde B, et al. The involvement of clathrin-mediated endocytosis and two Sid-1-like transmembrane proteins in double-stranded RNA uptake in the Colorado potato beetle midgut [J]. Insect Mol Biol, 2016, 25(3): 315-323. |
| [132] | Wang M, Weiberg A, Jin HL. Pathogen small RNAs: a new class of effectors for pathogen attacks [J]. Mol Plant Pathol, 2015, 16(3): 219-223. |
| [133] | Wytinck N, Sullivan DS, Biggar KT, et al. Clathrin mediated endocytosis is involved in the uptake of exogenous double-stranded RNA in the white mold phytopathogen Sclerotinia sclerotiorum [J]. Sci Rep, 2020, 10(1): 12773. |
| [134] | Dubelman S, Fischer J, Zapata F, et al. Environmental fate of double-stranded RNA in agricultural soils [J]. PLoS One, 2014, 9(3): e93155. |
| [135] | Mitter N, Worrall EA, Robinson KE, et al. Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses [J]. Nat Plants, 2017, 3: 16207. |
| [136] | Demirer GS, Zhang H, Goh NS, et al. Carbon nanocarriers deliver siRNA to intact plant cells for efficient gene knockdown [J]. Sci Adv, 2020, 6(26): eaaz0495. |
| [137] | Qiao LL, Niño-Sánchez J, Hamby R, et al. Artificial nanovesicles for dsRNA delivery in spray-induced gene silencing for crop protection [J]. Plant Biotechnol J, 2023, 21(4): 854-865. |
| [138] | Cai Q, He BY, Weiberg A, et al. Small RNAs and extracellular vesicles: new mechanisms of cross-species communication and innovative tools for disease control [J]. PLoS Pathog, 2019, 15(12): e1008090. |
| [139] | Luo XM, Nanda S, Zhang YJ, et al. Risk assessment of RNAi-based biopesticides [J]. New Crops, 2024, 1: 100019. |
| [140] | Hoang BTL, Fletcher SJ, Brosnan CA, et al. RNAi as a foliar spray: efficiency and challenges to field applications [J]. Int J Mol Sci, 2022, 23(12): 6639. |
| [141] | Zhu F, Xu JJ, Palli R, et al. Ingested RNA interference for managing the populations of the Colorado potato beetle, Leptinotarsa decemlineata [J]. Pest Manag Sci, 2011, 67(2): 175-182. |
| [142] | Rodrigues TB, Mishra SK, Sridharan K, et al. First sprayable double-stranded RNA-based biopesticide product targets Proteasome subunit beta type-5 in Colorado potato beetle (Leptinotarsa decemlineata) [J]. Front Plant Sci, 2021, 12: 728652. |
| [143] | McLoughlin AG, Wytinck N, Walker PL, et al. Identification and application of exogenous dsRNA confers plant protection against Sclerotinia sclerotiorum and Botrytis cinerea [J]. Sci Rep, 2018, 8(1): 7320. |
| [144] | Forster H, Shuai B. Exogenous siRNAs against chitin synthase gene suppress the growth of the pathogenic fungus Macrophomina phaseolina [J]. Mycologia, 2020, 112(4): 699-710. |
| [145] | Hu DF, Chen ZY, Zhang CQ, et al. Reduction of Phakopsora pachyrhizi infection on soybean through host- and spray-induced gene silencing [J]. Mol Plant Pathol, 2020, 21(6): 794-807. |
| [146] | Werner BT, Gaffar FY, Schuemann J, et al. RNA-spray-mediated silencing of Fusarium graminearum AGO and DCL genes improve barley disease resistance [J]. Front Plant Sci, 2020, 11: 476. |
| [147] | Konakalla NC, Kaldis A, Berbati M, et al. Exogenous application of double-stranded RNA molecules from TMV p126 and CP genes confers resistance against TMV in tobacco [J]. Planta, 2016, 244(4): 961-969. |
| [148] | Vadlamudi T, Patil BL, Kaldis A, et al. DsRNA-mediated protection against two isolates of Papaya ringspot virus through topical application of dsRNA in Papaya [J]. J Virol Meth, 2020, 275: 113750. |
| [25] | Kumagai MH, Donson J, della-Cioppa G, et al. Cytoplasmic inhibition of carotenoid biosynthesis with virus-derived RNA [J]. Proc Natl Acad Sci USA, 1995, 92(5): 1679-1683. |
| [26] | van Kammen A. Virus-induced gene silencing in infected and transgenic plants [J]. Trends Plant Sci, 1997, 2(11): 409-411. |
| [27] | Ruiz MT, Voinnet O, Baulcombe DC. Initiation and maintenance of virus-induced gene silencing [J]. Plant Cell, 1998, 10(6): 937-946. |
| [28] | Liu YL, Schiff M, Dinesh-Kumar SP. Virus-induced gene silencing in tomato [J]. Plant J, 2002, 31(6): 777-786. |
| [29] | Holzberg S, Brosio P, Gross C, et al. Barley stripe mosaic virus-induced gene silencing in a monocot plant [J]. Plant J, 2002, 30(3): 315-327. |
| [30] | Zhang CQ, Ghabrial SA. Development of Bean pod mottle virus-based vectors for stable protein expression and sequence-specific virus-induced gene silencing in soybean [J]. Virology, 2006, 344(2): 401-411. |
| [31] | Senthil-Kumar M, Mysore KS. New dimensions for VIGS in plant functional genomics [J]. Trends Plant Sci, 2011, 16(12): 656-665. |
| [32] | Kjemtrup S, Sampson KS, Peele CG, et al. Gene silencing from plant DNA carried by a geminivirus [J]. Plant J, 1998, 14(1): 91-100. |
| [33] | Turnage MA, Muangsan N, Peele CG, et al. Geminivirus-based vectors for gene silencing in Arabidopsis [J]. Plant J, 2002, 30(1): 107-114. |
| [34] | Gosselé V, Faché I, Meulewaeter F, et al. SVISS-a novel transient gene silencing system for gene function discovery and validation in tobacco plants [J]. Plant J, 2002, 32(5): 859-866. |
| [35] | Zhang Y, Xian YB, Yang H, et al. A novel geminivirus-derived 3' flanking sequence of terminator mediates the gene expression enhancement [J]. Plant Biotechnol J, 2025, 23(4): 1053-1066. |
| [36] | Zerbini FM, Briddon RW, Idris A, et al. ICTV virus taxonomy profile: Geminiviridae [J]. J Gen Virol, 2017, 98(2): 131-133. |
| [149] | Mumbanza FM, Kiggundu A, Tusiime G, et al. In vitro antifungal activity of synthetic dsRNA molecules against two pathogens of banana, Fusarium oxysporum f. sp. cubense and Mycosphaerella fijiensis [J]. Pest Manag Sci, 2013, 69(10): 1155-1162. |
| [150] | Gu KX, Song XS, Xiao XM, et al. A β2-tubulin dsRNA derived from Fusarium asiaticum confers plant resistance to multiple phytopathogens and reduces fungicide resistance [J]. Pestic Biochem Physiol, 2019, 153: 36-46. |
| [151] | Guan RB, Chu DD, Han XY, et al. Advances in the development of microbial double-stranded RNA production systems for application of RNA interference in agricultural pest control [J]. Front Bioeng Biotechnol, 2021, 9: 753790. |
| [152] | Rodrigues T, Sridharan K, Manley B, et al. Development of dsRNA as a sustainable bioinsecticide: from laboratory to field [M]//Crop Protection Products for Sustainable Agriculture. Washington, DC: American Chemical Society, 2021: 65-82. |
| [153] | Uslu VV, Bassler A, Krczal G, et al. High-pressure-sprayed double stranded RNA does not induce RNA interference of a reporter gene [J]. Front Plant Sci, 2020, 11: 534391. |
| [154] | Elston KM, Maeda GP, Perreau J, et al. Addressing the challenges of symbiont-mediated RNAi in aphids [J]. PeerJ, 2023, 11: e14961. |
| [155] | Horn T, Boutros M. E-RNAi: a web application for the multi-species design of RNAi reagents—2010 update [J]. Nucleic Acids Res, 2010, 38(Web Server issue): W332-W339. |
| [156] | Cui CL, Wang Y, Li YF, et al. Expression of mosquito miRNAs in entomopathogenic fungus induces pathogen-mediated host RNA interference and increases fungal efficacy [J]. Cell Rep, 2022, 41(4): 111527. |
| [157] | Lin SJ, Zhang Q, Bai SY, et al. Beyond species and spatial boundaries: enabling long-distance gene silencing in plants via guanidinium-siRNA nanoparticles [J]. Plant Biotechnol J, 2025, 23(4): 1165-1177. |
| [158] | Yu HP, Yang H, Sun WQ, et al. An interpretable RNA foundation model for exploring functional RNA motifs in plants [J]. Nat Mach Intell, 2024, 6(12): 1616-1625. |
| [159] | Hou X, He Y, Fang P, et al. Using artificial intelligence to document the hidden RNA virosphere [J]. Cell, 2024, 187(24): 6929-6942.e16. |
| [37] | Hanley-Bowdoin L, Bejarano ER, Robertson D, et al. Geminiviruses: Masters at redirecting and reprogramming plant processes [J]. Nat Rev Microbiol, 2013, 11(11): 777-788. |
| [38] | Blevins T, Rajeswaran R, Aregger M, et al. Massive production of small RNAs from a non-coding region of Cauliflower mosaic virus in plant defense and viral counter-defense [J]. Nucleic Acids Res, 2011, 39(12): 5003-5014. |
| [39] | Nagy PD, Pogany J. Yeast as a model host to dissect functions of viral and host factors in tombusvirus replication [J]. Virology, 2006, 344(1): 211-220. |
| [40] | Dreher TW, Miller WA. Translational control in positive strand RNA plant viruses [J]. Virology, 2006, 344(1): 185-197. |
| [41] | Kushner DB, Lindenbach BD, Grdzelishvili VZ, et al. Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus [J]. Proc Natl Acad Sci USA, 2003, 100(26): 15764-15769. |
| [42] | Fukudome A, Fukuhara T. Plant dicer-like proteins: double-stranded RNA-cleaving enzymes for small RNA biogenesis [J]. J Plant Res, 2017, 130(1): 33-44. |
| [43] | Thomas CL, Jones L, Baulcombe DC, et al. Size constraints for targeting post-transcriptional gene silencing and for RNA-directed methylation in Nicotiana benthamiana using a potato virus X vector [J]. Plant J, 2001, 25(4): 417-425. |
| [44] | Fantini E, Giuliano G. Virus-induced gene silencing as a tool to study tomato fruit biochemistry [J]. Methods Mol Biol, 2016, 1363: 65-78. |
| [45] | Jacob SS, Vanitharani R, Karthikeyan AS, et al. Mungbean yellow mosaic virus-Vi agroinfection by codelivery of DNA A and DNA B from one Agrobacterium strain [J]. Plant Dis, 2003, 87(3): 247-251. |
| [46] | Lee WS, Rudd JJ, Kanyuka K. Virus induced gene silencing (VIGS) for functional analysis of wheat genes involved in Zymoseptoria tritici susceptibility and resistance [J]. Fungal Genet Biol, 2015, 79: 84-88. |
| [47] | Pflieger S, Blanchet S, Camborde L, et al. Efficient virus-induced gene silencing in Arabidopsis using a ‘one-step’ TYMV-derived vector [J]. Plant J, 2008, 56(4): 678-690. |
| [48] | Yuan WM, Li Y, Zhang WJ, et al. Pinpointing MQTLs and candidate genes related to early maturity in upland cotton through the integration of meta-analysis, RNA-seq, and VIGS approaches [J]. Ind Crops Prod, 2025, 223: 120195. |
| [49] | Hands P, Vosnakis N, Betts D, et al. Alternate transcripts of a floral developmental regulator have both distinct and redundant functions in opium poppy [J]. Ann Bot, 2011, 107(9): 1557-1566. |
| [50] | Liang YW, Gao Q, Li F, et al. The giant genome of lily provides insights into the hybridization of cultivated lilies [J]. Nat Commun, 2025, 16(1): 45. |
| [51] | Zhang YQ, Zhang XW, Yu L, et al. An efficient rice virus-induced gene silencing system mediated by wheat dwarf virus [J]. Appl Sci, 2025, 15(11): 5818. |
| [52] | Kolodziej MC, Singla J, Sánchez-Martín J, et al. A membrane-bound ankyrin repeat protein confers race-specific leaf rust disease resistance in wheat [J]. Nat Commun, 2021, 12(1): 956. |
| [53] | Murphree C, Kim SB, Karre S, et al. Use of virus-induced gene silencing to characterize genes involved in modulating hypersensitive cell death in maize [J]. Mol Plant Pathol, 2020, 21(12): 1662-1676. |
| [54] | He XY, Zeng JB, Cao FB, et al. HvEXPB7, a novel β-expansin gene revealed by the root hair transcriptome of Tibetan wild barley, improves root hair growth under drought stress [J]. J Exp Bot, 2015, 66(22): 7405-7419. |
| [55] | Zhai N, Jia HH, Liu DD, et al. GhMAP3K65 a cotton raf-like MAP3K gene, enhances susceptibility to pathogen infection and heat stress by negatively modulating growth and development in transgenic Nicotiana benthamiana [J]. Int J Mol Sci, 2017, 18(11): 2462. |
| [56] | O'Rourke JA, Graham MA. Coupling VIGS with short- and long-term stress exposure to understand the fiskeby III iron deficiency stress response [J]. Int J Mol Sci, 2022, 24(1): 647. |
| [57] | Fei LY, Liu JR, Liao Y, et al. The CaABCG14 transporter gene regulates the capsaicin accumulation in pepper septum [J]. Int J Biol Macromol, 2024, 280(Pt 4): 136122. |
| [58] | Azeez A, Bates PD. Self-incompatibility based functional genomics for rapid phenotypic characterization of seed metabolism genes [J]. Plant Biotechnol J, 2024, 22(10): 2688-2690. |
| [59] | Chen YS, Li Y, Luo GQ, et al. Gene identification, expression analysis, and molecular docking of SAT and OASTL in the metabolic pathway of selenium in Cardamine hupingshanensis [J]. Plant Cell Rep, 2024, 43(6): 148. |
| [60] | Cheng LN, Li RZ, Wang XY, et al. A SlCLV3-SlWUS module regulates auxin and ethylene homeostasis in low light-induced tomato flower abscission [J]. Plant Cell, 2022, 34(11): 4388-4408. |
| [61] | Geng ZK, Ma L, Rong YL, et al. A hydrogen-sulfide-repressed methionine synthase SlMS1 acts as a positive regulator for fruit ripening in tomato [J]. Int J Mol Sci, 2022, 23(20): 12239. |
| [62] | Alvarez JP, Pekker I, Goldshmidt A, et al. Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species [J]. Plant Cell, 2006, 18(5): 1134-1151. |
| [63] | Yan J, Gu YY, Jia XY, et al. Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis [J]. Plant Cell, 2012, 24(2): 415-427. |
| [64] | Felippes FF, Weigel D. Triggering the formation of tasiRNAs in Arabidopsis thaliana: the role of microRNA miR173 [J]. EMBO Rep, 2009, 10(3): 264-270. |
| [65] | Tang Y, Wang F, Zhao JP, et al. Virus-based microRNA expression for gene functional analysis in plants[J]. Plant Physiol, 2010, 153(2): 632-641. |
| [66] | Akgul B, Aydinoglu F. Evaluation of zma-miR408 and its target genes function on maize (Zea mays) leaf growth response to cold stress by VIGS-based STTM approach [J]. Gene, 2025, 938: 149161. |
| [67] | Zhan JJ, Chu Y, Wang Y, et al. The miR164-GhCUC2-GhBRC1 module regulates plant architecture through abscisic acid in cotton [J]. Plant Biotechnol J, 2021, 19(9): 1839-1851. |
| [68] | Senthil-Kumar M, Mysore KS. Virus-induced gene silencing can persist for more than 2-years and also be transmitted to progeny seedlings in Nicotiana benthamiana and tomato [J]. Plant Biotechnol J, 2011, 9(7): 797-806. |
| [69] | Zulfiqar S, Farooq MA, Zhao TT, et al. Virus-induced gene silencing (VIGS): a powerful tool for crop improvement and its advancement towards epigenetics [J]. Int J Mol Sci, 2023, 24(6): 5608. |
| [70] | Wang CQ, Ma ZM, Zhou JH, et al. A simple and effective VIGS system facilitates the control of Citrus canker by silencing CsLOB1 [J]. Phytopathol Res, 2024, 6(1): 12. |
| [71] | Ahn CS, Pai HS. Physiological function of IspE, a plastid MEP pathway gene for isoprenoid biosynthesis, in organelle biogenesis and cell morphogenesis in Nicotiana benthamiana [J]. Plant Mol Biol, 2008, 66(5): 503-517. |
| [72] | Gao W, Long L, Zhu LF, et al. Proteomic and virus-induced gene silencing (VIGS) analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae [J]. Mol Cell Proteom, 2013, 12(12): 3690-3703. |
| [73] | Bennypaul HS, Mutti JS, Rustgi S, et al. Virus-induced gene silencing (VIGS) of genes expressed in root, leaf, and meiotic tissues of wheat [J]. Funct Integr Genomics, 2012, 12(1): 143-156. |
| [74] | Sahu PP, Puranik S, Khan M, et al. Recent advances in tomato functional genomics: utilization of VIGS [J]. Protoplasma, 2012, 249(4): 1017-1027. |
| [75] | Unver T, Budak H. Virus-induced gene silencing, a post transcriptional gene silencing method [J]. Int J Plant Genom, 2009, 2009(1): 198680. |
| [76] | Rössner C, Lotz D, Becker A. VIGS goes viral: how VIGS transforms our understanding of plant science [J]. Annu Rev Plant Biol, 2022, 73: 703-728. |
| [77] | Reyes MI, Flores-Vergara MA, Guerra-Peraza O, et al. A VIGS screen identifies immunity in the Arabidopsis Pla-1 accession to viruses in two different genera of the Geminiviridae [J]. Plant J, 2017, 92(5): 796-807. |
| [78] | Tian J, Pei HX, Zhang S, et al. TRV-GFP a modified Tobacco rattle virus vector for efficient and visualizable analysis of gene function [J]. J Exp Bot, 2014, 65(1): 311-322. |
| [79] | Boutla A, Kalantidis K, Tavernarakis N, et al. Induction of RNA interference in Caenorhabditis elegans by RNAs derived from plants exhibiting post-transcriptional gene silencing [J]. Nucleic Acids Res, 2002, 30(7): 1688-1694. |
| [80] | Huang GZ, Allen R, Davis EL, et al. Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene [J]. Proc Natl Acad Sci USA, 2006, 103(39): 14302-14306. |
| [81] | Nowara D, Gay A, Lacomme C, et al. HIGS host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis [J]. Plant Cell, 2010, 22(9): 3130-3141. |
| [82] | Weiberg A, Wang M, Lin FM, et al. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways [J]. Science, 2013, 342(6154): 118-123. |
| [83] | Zhang T, Zhao YL, Zhao JH, et al. Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen [J]. Nat Plants, 2016, 2(10): 16153. |
| [84] | Xiao D, Gao X, Xu J, et al. Clathrin-dependent endocytosis plays a predominant role in cellular uptake of double-stranded RNA in the red flour beetle [J]. Insect Biochem Mol Biol, 2015, 60: 68-77. |
| [85] | Winston WM, Molodowitch C, Hunter CP. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1 [J]. Science, 2002, 295(5564): 2456-2459. |
| [86] | Zhang J, Khan SA, Hasse C, et al. Pest control. Full crop protection from an insect pest by expression of long double-stranded RNAs in plastids [J]. Science, 2015, 347(6225): 991-994. |
| [87] | Zand Karimi H, Innes RW. Molecular mechanisms underlying host-induced gene silencing [J]. Plant Cell, 2022, 34(9): 3183-3199. |
| [88] | Biedenkopf D, Will T, Knauer T, et al. Systemic spreading of exogenous applied RNA biopesticides in the crop plant Hordeum vulgare [J]. ExRNA, 2020, 2: 12. |
| [1] | LAI Shi-yu, LIANG Qiao-lan, WEI Lie-xin, NIU Er-bo, CHEN Ying-e, ZHOU Xin, YANG Si-zheng, WANG Bo. The Role of NbJAZ3 in the Infection of Nicotiana benthamiana by Alfalfa Mosaic Virus [J]. Biotechnology Bulletin, 2025, 41(8): 186-196. |
| [2] | WEN Jing, LI Qian-qian, ZHANG Ming-da, TAN Ming-yue, JIN Bo-yang, SHEN XIU-li, DU Zhi-qiang. Molecular Mechanism of Duox 2 Regulating Innate Immunity against Bacteria in Procambarus clarkii Intestine [J]. Biotechnology Bulletin, 2025, 41(1): 324-332. |
| [3] | WANG Ke-ran, YAN Jun-jie, LIU Jian-feng, GAO Yu-lin. Application and Risk of RNAi Technology in Potato Insect Pest Management [J]. Biotechnology Bulletin, 2024, 40(9): 4-10. |
| [4] | LI Yi-jun, YANG Xiao-bei, XIA Lin, LUO Zhao-peng, XU Xin, YANG Jun, NING Qian-ji, WU Ming-zhu. Cloning and Functional Analysis of NtPRR37 Gene in Nicotiana tabacum L. [J]. Biotechnology Bulletin, 2024, 40(8): 221-231. |
| [5] | JIN Bo-yang, QIN Shi-yu, ZHANG Ming-da, LI Qian-qian, WEN Jing, SHEN Xiu-li, DU Zhi-qiang. Research on the Molecular Mechanism of Crayfish prx 6 in the Process of Defending against Staphylococcus aureus Infection [J]. Biotechnology Bulletin, 2024, 40(7): 314-322. |
| [6] | HUA Xuan, TIAN Bo-wen, ZHOU Xin-tong, JIANG Zi-han, WANG Shi-qi, HUANG Qian-hui, ZHANG Jian, CHEN Yan-hong. Cloning SmERF B3-45 from Salix matsudana and Functional Analysis on Its Tolerance to Salt [J]. Biotechnology Bulletin, 2024, 40(12): 124-135. |
| [7] | ZHAO Jian-hua, GAO Feng, LIU Qing-yan, GUO Hui-shan. RNA Silencing Efficiency Affected by RNA Structure [J]. Biotechnology Bulletin, 2024, 40(10): 19-29. |
| [8] | ZHANG Yi, ZHANG Xin-ru, ZHANG Jin-ke, HU Li-zong, SHANGGUAN Xin-xin, ZHENG Xiao-hong, HU Juan-juan, ZHANG Cong-cong, MU Gui-qing, LI Cheng-wei. Functional Analysis of TaMYB1 Gene in Wheat Under Cadmium Stress [J]. Biotechnology Bulletin, 2024, 40(1): 194-206. |
| [9] | LIU Zhen-yin, DUAN Zhi-zhen, PENG Ting, WANG Tong-xin, WANG Jian. Establishment and Optimization of Virus-induced Gene Silencing System in Bougainvillea peruviana ‘Thimma’ [J]. Biotechnology Bulletin, 2023, 39(7): 123-130. |
| [10] | LI Wen-chen, LIU Xin, KANG Yue, LI Wei, QI Ze-zheng, YU Lu, WANG Fang. Optimization and Application of Tobacco Rattle Virus-induced Gene Silencing System in Soybean [J]. Biotechnology Bulletin, 2023, 39(7): 143-150. |
| [11] | ZHANG Long-xi, LYU Lin, ZHANG Huan-huan, ZHOU Jin-cheng, CHE Wu-nan, DONG Hui. Research Progress in the Application of RNAi Technology in Parasitoid Wasps [J]. Biotechnology Bulletin, 2023, 39(12): 99-108. |
| [12] | LI Xiu-qing, HU Zi-yao, LEI Jian-feng, DAI Pei-hong, LIU Chao, DENG Jia-hui, LIU Min, SUN Ling, LIU Xiao-dong, LI Yue. Cloning and Functional Analysis of Gene GhTIFY9 Related to Cotton Verticillium Wilt Resistance [J]. Biotechnology Bulletin, 2022, 38(8): 127-134. |
| [13] | FU Si-tong, SI Wei-jia, LIU Ying, CHENG Tang-ren, WANG Jia, ZHANG Qi-xiang, PAN Hui-tang. Establishing Tobacco Rattle Virus-mediated Gene Silencing System for Primula forbesii [J]. Biotechnology Bulletin, 2022, 38(4): 295-302. |
| [14] | LIU Xiao-mei, WANG Dong-xin, ZHANG Chun, WEI Shuang-shi. Inhibition of AAV-mediated RNAi to SARS-CoV-2 S Gene Expression [J]. Biotechnology Bulletin, 2022, 38(3): 188-193. |
| [15] | GUO Yu-fei, YAN Rong-mei, ZHANG Xiao-ru, CAO Wei, LIU Hao. Metabolic Engineering Modification of Aspergillus niger for the Production of D-glucaric Acid [J]. Biotechnology Bulletin, 2022, 38(11): 227-237. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||