








Biotechnology Bulletin ›› 2026, Vol. 42 ›› Issue (7): 1-13.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0964
GAO Fei, ZHANG Yu-xi, MU Di, CHEN Zheng, CHEN Hong-yan( )
)
Received:2025-09-09
Online:2026-02-09
Published:2026-02-09
Contact:
CHEN Hong-yan
E-mail:03692@qqhru.edu.cn
GAO Fei, ZHANG Yu-xi, MU Di, CHEN Zheng, CHEN Hong-yan. Isolation, Identification, and Whole-genome Sequencing Analysis of Murine-Derived Limosilactobacillus reuteri[J]. Biotechnology Bulletin, 2026, 42(7): 1-13.
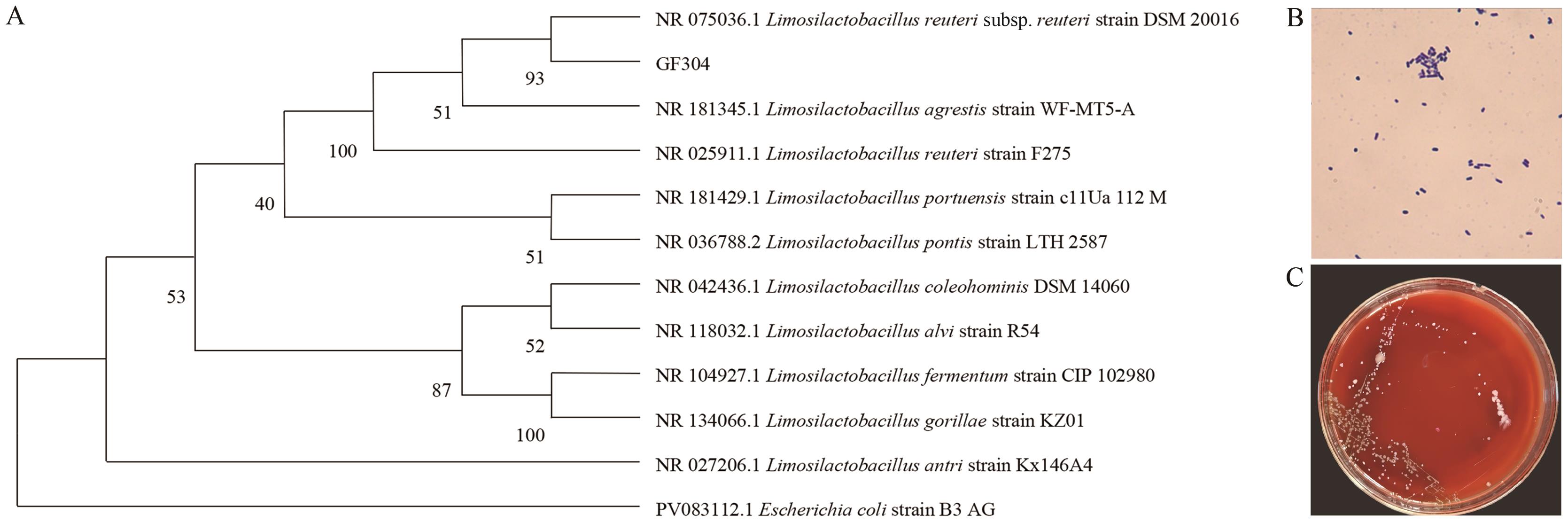
Fig. 1 Morphological observation, hemolytic assay, and phylogenetic tree of the isolated strainsA: Gram staining microscopy (×1 000). B: Hemolysis test results of strain GF304. C: Phylogenetic tree of strain GF304
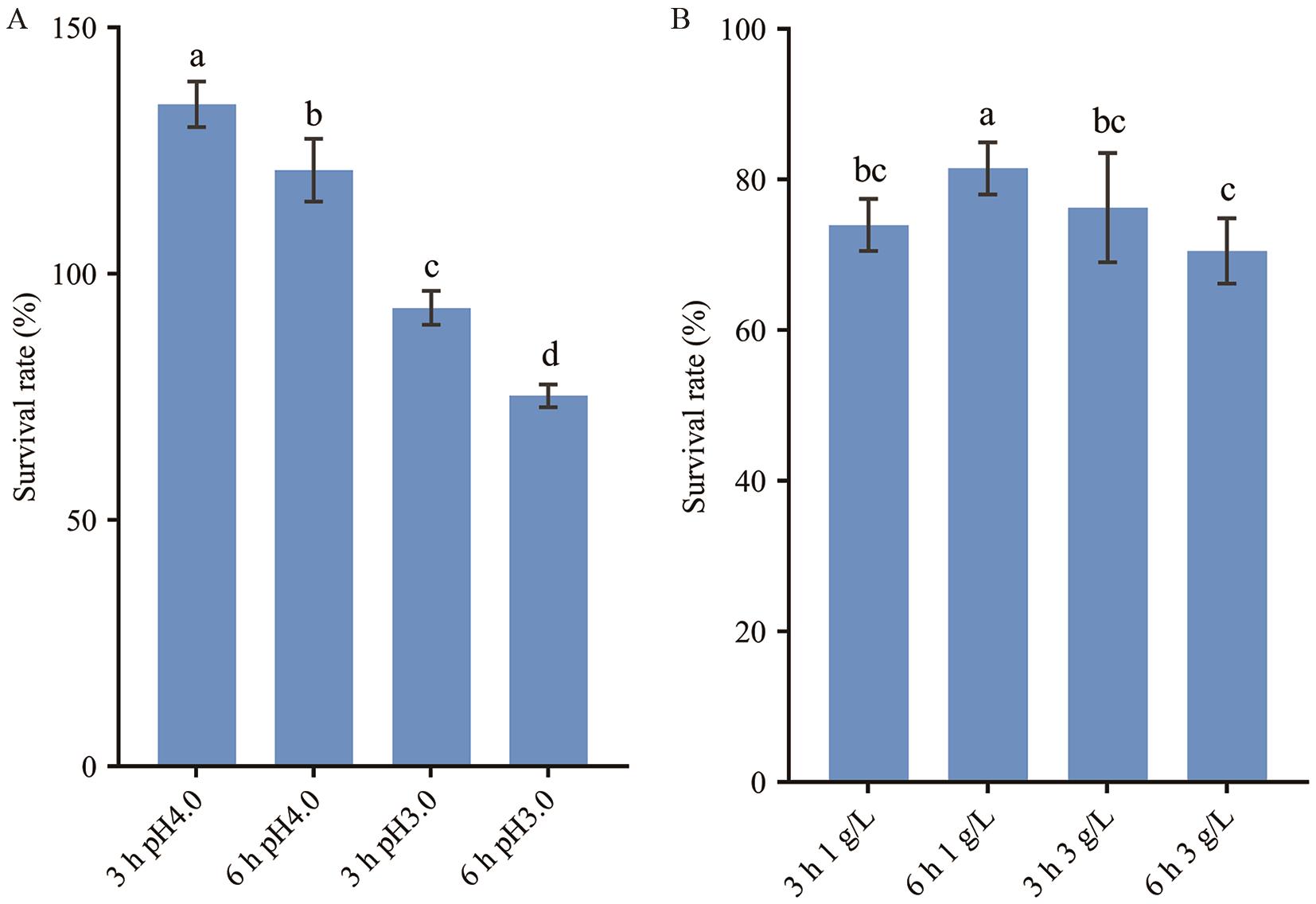
Fig. 3 Tolerances to acid and choline of L. reuteri GF304A: Tolerance to acid. B: Tolerance to choline. Different lowercase letters indicate significant differences between groups (P<0.05)
| 菌株 | 人工胃液 | 人工肠液 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 活菌数(×106 CFU·mL-1) | 存活率(%) | 活菌数(×105 CFU·mL-1) | 存活率/% | ||||||
| 0 h | 2 h | 2 h/0 h | 0 h | 2 h | 4 h | 2 h/0 h | 4 h/0 h | ||
| GF304 | 3.26±0.16 | 2.73±0.43 | 83.38±0.09 | 4.75±0.67 | 4.00±0.85 | 2.72±0.70 | 83.66±0.07 | 57.80±0.14 | |
Table 1 Survival rate of L. reuteri GF304 in simulated gastric juice and simulated intestinal juice
| 菌株 | 人工胃液 | 人工肠液 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 活菌数(×106 CFU·mL-1) | 存活率(%) | 活菌数(×105 CFU·mL-1) | 存活率/% | ||||||
| 0 h | 2 h | 2 h/0 h | 0 h | 2 h | 4 h | 2 h/0 h | 4 h/0 h | ||
| GF304 | 3.26±0.16 | 2.73±0.43 | 83.38±0.09 | 4.75±0.67 | 4.00±0.85 | 2.72±0.70 | 83.66±0.07 | 57.80±0.14 | |
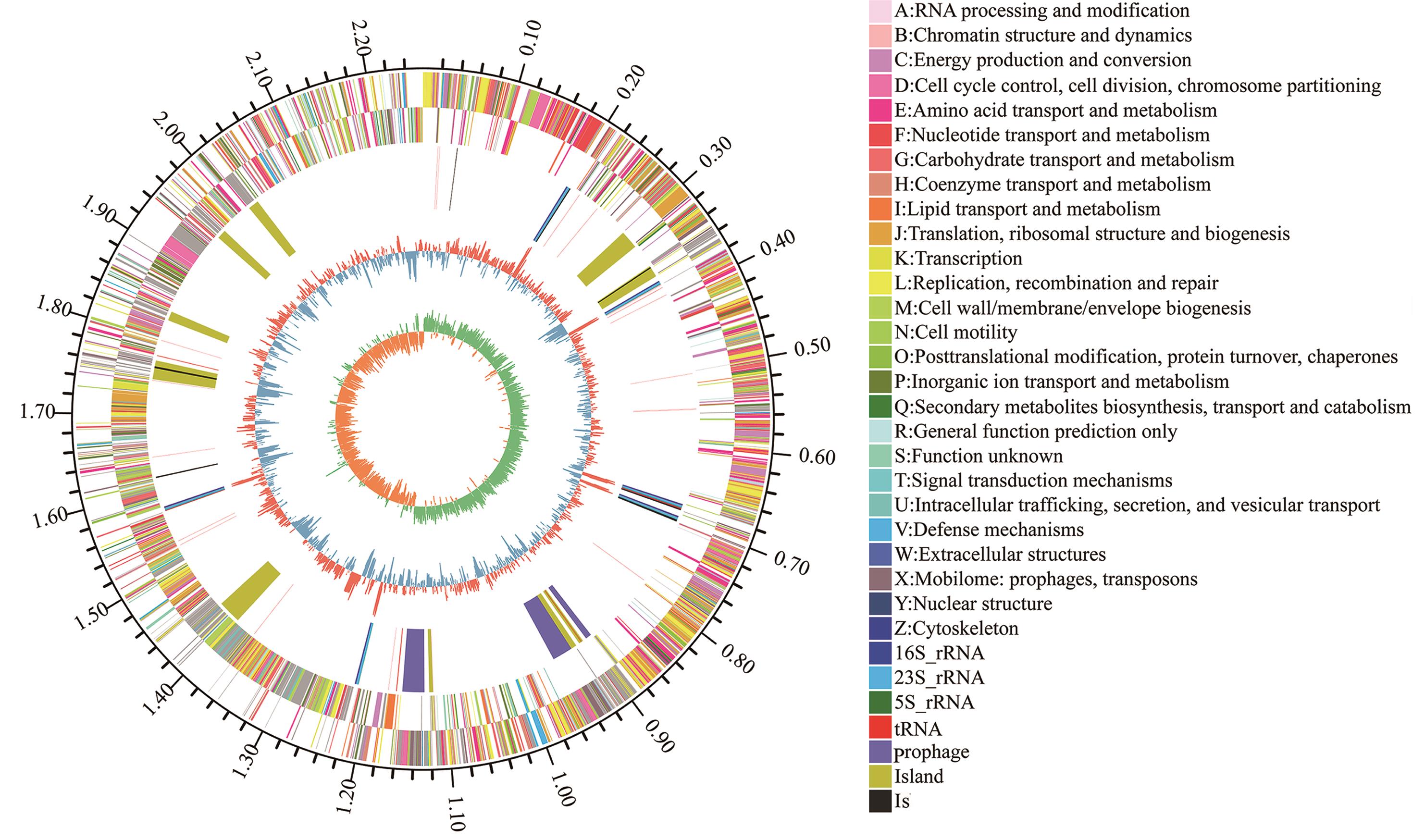
Fig. 5 The circular genome map of L. reuteri strain GF304From the outermost to the innermost layer, the concentric rings represent: the genome scale marker; coding sequences (CDSs) on the positive and negative strands annotated according to COG functional classifications; the distribution of rRNAs and tRNAs; GC content (red outward peaks indicate higher than the average genomic GC content, while blue inward peaks indicate lower than the average); and the innermost layer indicates GC skew (green indicates G > C, yellow indicates C > G)
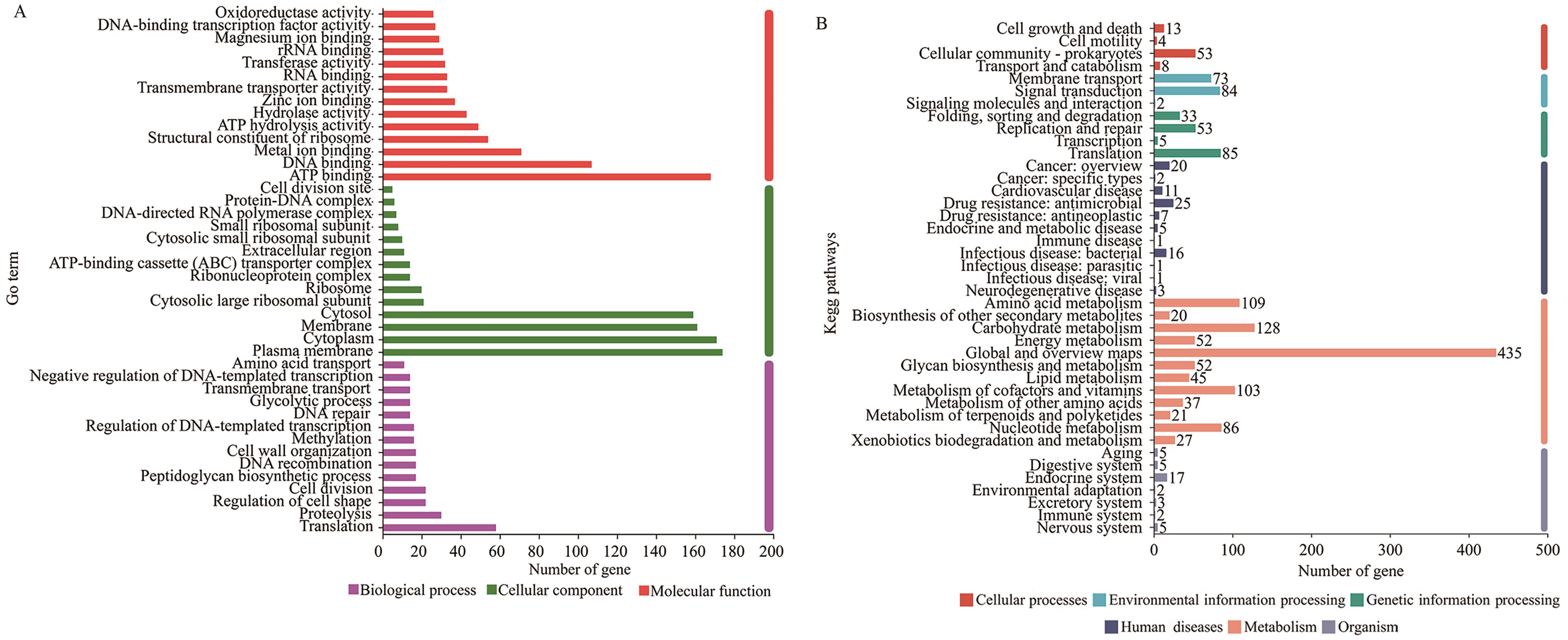
Fig. 6 GO functional classification and KEGG metabolic pathway classification of the genome of L. reuteri strain GF304A: GO Analysis. B: KEGG Analysis
| Gene name | Gene product | Gene ID |
|---|---|---|
| Temperature | ||
| groES | Co-chaperone GroES | gene0399 |
| groEL | Chaperonin GroEL | gene0400 |
| smpB | SsrA-binding protein SmpB | gene0450 |
| hrcA | Heat-inducible transcriptional repressor HrcA | gene0768 |
| dnaK | Molecular chaperone DnaK | gene0770 |
| dnaJ | Molecular chaperone DnaJ | gene0771 |
| cspA | Cold shock domain-containing protein,cold-shock protein | gene0676/gene1783 |
| pH | ||
| atpB | F0F1 ATP synthase subunit A | gene0512 |
| atpE | F0F1 ATP synthase subunit C | gene0513 |
| atpF | F0F1 ATP synthase subunit B | gene0514 |
| atpH | ATP synthase F1 subunit delta | gene0515 |
| atpA | F0F1 ATP synthase subunit alpha | gene0516 |
| atpG | F0F1 ATP synthase subunit gamma | gene0517 |
| atpD | F0F1 ATP synthase subunit beta | gene0518 |
| atpC | F0F1 ATP synthase subunit epsilon | gene0519 |
| - | Alkaline shock response membrane anchor protein AmaP | gene0966 |
| arcA | Arginine deiminase | gene0496 |
| arcC | Carbamate kinase | gene0480 |
| argR | Arginine repressor/ArgR family transcriptional regulator | gene0497/gene1329 |
| argG | Argininosuccinate synthase | gene0802 |
| argH | Argininosuccinate lyase | gene0803 |
| gadB | Glutamate decarboxylase | gene0539 |
| gadC | Gamma-aminobutyrate antiporte/glutamate/gamma-aminobutyrate family transporter YjeM | gene0542/gene0540/gene1458/gene1618 |
| nhaC | Na+/H+ antiporter NhaC family protein/Na+/H+ antiporter NhaC | gene0171/gene2094 |
| Bile salt resistance | ||
| cbh | Choloylglycine hydrolase | gene0801 |
| ppaC | Manganese-dependent inorganic pyrophosphatase | gene0959 |
| Production of adhesion molecules | ||
| ltaS | LTA synthase family protein | gene1935/gene2098 |
| Oxidative resistance | ||
| trxA | Thioredoxin | gene0582/gene2088 |
| trxB | Thioredoxin-disulfide reductase | gene0423 |
| gshA | Glutamate--cysteine ligase | gene0072/gene1632 |
| pepN | M1 family metallopeptidase | gene2145 |
| nfrA1 | NADPH-dependent oxidoreductase | gene1863 |
| Riboflavin biosynthesis | ||
| ribD | Bifunctional diaminohydroxyphosphoribosylaminopyrimidine Deaminase/5-amino-6-(5-phosphoribosylamino)uracil reductase RibD | gene0993 |
| ribE | riboflavin synthase | gene0994 |
| ribBA | Bifunctional 3,4-dihydroxy-2-butanone-4-phosphate synthase/GTP cyclohydrolase Ⅱ | gene0995 |
| ribH | 6,7-dimethyl-8-ribityllumazine synthase | gene0996 |
| ribF | Riboflavin biosynthesis protein RibF | gene0766 |
| ribT | Reductase | gene0825 |
| Organic acid biosynthesis | ||
| ackA | Acetate kinase | gene0604 |
Table 2 Genes related to probiotic properties in Limosilactobacillus reuteri strain GF304
| Gene name | Gene product | Gene ID |
|---|---|---|
| Temperature | ||
| groES | Co-chaperone GroES | gene0399 |
| groEL | Chaperonin GroEL | gene0400 |
| smpB | SsrA-binding protein SmpB | gene0450 |
| hrcA | Heat-inducible transcriptional repressor HrcA | gene0768 |
| dnaK | Molecular chaperone DnaK | gene0770 |
| dnaJ | Molecular chaperone DnaJ | gene0771 |
| cspA | Cold shock domain-containing protein,cold-shock protein | gene0676/gene1783 |
| pH | ||
| atpB | F0F1 ATP synthase subunit A | gene0512 |
| atpE | F0F1 ATP synthase subunit C | gene0513 |
| atpF | F0F1 ATP synthase subunit B | gene0514 |
| atpH | ATP synthase F1 subunit delta | gene0515 |
| atpA | F0F1 ATP synthase subunit alpha | gene0516 |
| atpG | F0F1 ATP synthase subunit gamma | gene0517 |
| atpD | F0F1 ATP synthase subunit beta | gene0518 |
| atpC | F0F1 ATP synthase subunit epsilon | gene0519 |
| - | Alkaline shock response membrane anchor protein AmaP | gene0966 |
| arcA | Arginine deiminase | gene0496 |
| arcC | Carbamate kinase | gene0480 |
| argR | Arginine repressor/ArgR family transcriptional regulator | gene0497/gene1329 |
| argG | Argininosuccinate synthase | gene0802 |
| argH | Argininosuccinate lyase | gene0803 |
| gadB | Glutamate decarboxylase | gene0539 |
| gadC | Gamma-aminobutyrate antiporte/glutamate/gamma-aminobutyrate family transporter YjeM | gene0542/gene0540/gene1458/gene1618 |
| nhaC | Na+/H+ antiporter NhaC family protein/Na+/H+ antiporter NhaC | gene0171/gene2094 |
| Bile salt resistance | ||
| cbh | Choloylglycine hydrolase | gene0801 |
| ppaC | Manganese-dependent inorganic pyrophosphatase | gene0959 |
| Production of adhesion molecules | ||
| ltaS | LTA synthase family protein | gene1935/gene2098 |
| Oxidative resistance | ||
| trxA | Thioredoxin | gene0582/gene2088 |
| trxB | Thioredoxin-disulfide reductase | gene0423 |
| gshA | Glutamate--cysteine ligase | gene0072/gene1632 |
| pepN | M1 family metallopeptidase | gene2145 |
| nfrA1 | NADPH-dependent oxidoreductase | gene1863 |
| Riboflavin biosynthesis | ||
| ribD | Bifunctional diaminohydroxyphosphoribosylaminopyrimidine Deaminase/5-amino-6-(5-phosphoribosylamino)uracil reductase RibD | gene0993 |
| ribE | riboflavin synthase | gene0994 |
| ribBA | Bifunctional 3,4-dihydroxy-2-butanone-4-phosphate synthase/GTP cyclohydrolase Ⅱ | gene0995 |
| ribH | 6,7-dimethyl-8-ribityllumazine synthase | gene0996 |
| ribF | Riboflavin biosynthesis protein RibF | gene0766 |
| ribT | Reductase | gene0825 |
| Organic acid biosynthesis | ||
| ackA | Acetate kinase | gene0604 |
Fig. 7 Analysis of secondary metabolite biosynthetic gene clusters in L. reuteri strain GF304A: T3PKS biosynthetic gene cluster. B: RiPP-like biosynthetic gene cluster
| [1] | Afrc RF. Probiotics in man and animals [J]. J Appl Bacteriol, 1989, 66(5): 365-378. |
| [2] | Lata P, Savitri. Probiotics and human health [J]. Res J Biotech, 2023, 18(7): 173-180. |
| [3] | Cristofori F, Dargenio VN, Dargenio C, et al. Anti-inflammatory and immunomodulatory effects of probiotics in gut inflammation: a door to the body [J]. Front Immunol, 2021, 12: 578386. |
| [4] | 程雨, 谢坤, 姜艳平, 等. 兔源罗伊乳杆菌的分离鉴定及益生效果评价 [J]. 中国兽医学报, 2024, 44(10): 2136-2144, 2293. |
| Cheng Y, Xie K, Jiang YP, et al. Isolation and identification of rabbit-derived Lactobacillus reuteri and evaluation of its probiotic function [J]. Chin J Vet Sci, 2024, 44(10): 2136-2144, 2293. | |
| [5] | Falcinelli S, Rodiles A, Hatef A, et al. Influence of probiotics administration on gut microbiota core: a review on the effects on appetite control, glucose, and lipid metabolism [J]. J Clin Gastroenterol, 2018, 52(): S50-S56. |
| [6] | 徐乐, 陈诗宇, 王上, 等. 茶花鸡源罗伊特氏黏液乳杆菌CHF7-2的益生特性及全基因组测序分析 [J]. 微生物学报, 2025, 65(3): 1197-1218. |
| Xu L, Chen SY, Wang S, et al. Probiotic characterization and whole genome sequencing of Limosilactobacillus reuteri CHF7-2 from Chahua chicken [J]. Acta Microbiol Sin, 2025, 65(3): 1197-1218. | |
| [7] | 国家卫生健康委员会. 解读关于《可用于食品的菌种名单》和《可用于婴幼儿食品的菌种名单》更新的公告(2022年第4号) [J]. 饮料工业, 2022, 25(5): 3-4. |
| National Health Commission of the People’s Republic of China. Interpretation of the announcement on updating the list of bacteria used in food and the list of bacteria used in infant food (No.4, 2022) [J]. Beverage Ind, 2022, 25(5): 3-4. | |
| [8] | Abuqwider J, Altamimi M, Mauriello G. Limosilactobacillus reuteri in health and disease [J]. Microorganisms, 2022, 10(3): 522. |
| [9] | 张晓莉, 杨勇, 王光强, 等. 基于全基因组和比较基因组分析罗伊氏粘液乳杆菌的益生特性 [J]. 中国食品学报, 2025, 25(8): 8-20. |
| Zhang XL, Yang Y, Wang GQ, et al. Probiotic properties of Limosilactobacillus reuteri based on whole-genome and comparative genome analysis [J]. J Chin Inst Food Sci Technol, 2025, 25(8): 8-20. | |
| [10] | 张媛媛, 赵梦迪, 李悦垚, 等. 犬源罗伊氏乳杆菌LRA7对比格犬短链脂肪酸含量、肠道菌群及代谢组的影响 [J]. 动物营养学报, 2025, 37(4): 2648-2660. |
| Zhang YY, Zhao MD, Li YY, et al. Effects of canine-derived Lactobacillus reuteri LRA7 on short-chain fatty acid contents, intestinal flora and metabolome in beagle dogs [J]. Chin J Anim Nutr, 2025, 37(4): 2648-2660. | |
| [11] | 刘春艳. 猪源罗伊氏乳杆菌对断奶仔猪肠道屏障功能和细胞外基质的影响 [D]. 南宁: 广西大学, 2023. |
| Liu CY. Effects of Lactobacillus reuteri from pigs on intestinal barrier function and extracellular matrix of weaned piglets [D]. Nanning: Guangxi University, 2023. | |
| [12] | 杨巍巍, 彭子维, 谢昊炅, 等. 罗伊氏黏液乳杆菌的分离鉴定及对瘤胃体外发酵参数的影响 [J]. 饲料工业, 2025, 46(15): 143-150. |
| Yang WW, Peng ZW, Xie HJ, et al. Isolation and identification of Limosilactobacillus reuteri and its effects on rumen fermentation parameters in vitro [J]. Feed Ind, 2025, 46(15): 143-150. | |
| [13] | 王志刚, 徐伟慧, 张迎, 等. 一种谷氨酸浓缩母液的常温脱盐以及资源化利用方法及其装置: CN115872482B [P]. 2023-08-11. |
| Wang ZG, Xu WH, Zhang Y, et al. A method and apparatus for room temperature desalination and resource utilization of glutamic acid concentrate mother liquor: CN115872482B [P]. 2023-08-11. | |
| [14] | 农业农村部办公厅关于印发《直接饲喂微生物和发酵制品生产菌株鉴定及其安全性评价指南》的通知(农办牧[2021]43号) [J]. 中华人民共和国农业农村部公报, 2021(11): 97-111. |
| Circular of the general office of the ministry of agriculture and rural affairs on printing and distributing the guidelines on identification and safety evaluation of direct-fed microbials and fermented-food-derived bacterial strains [J]. Gaz Minist Agric Rural Aff People’s Repub China, 2021(11): 97-111. | |
| [15] | Zhou WQ, Gao S, Zheng J, et al. Identification of an Aerococcus urinaeequi isolate by whole genome sequencing and average nucleotide identity analysis [J]. J Glob Antimicrob Resist, 2022, 29: 353-359. |
| [16] | 李兰溪. 植物乳杆菌LPJZ-658对成犬血液指标、消化吸收及肠道菌群的影响 [D]. 长春: 吉林农业大学, 2024. |
| Li LX. Effects of Lactiplantibacillus plantarum LPJZ-658 on blood indexes, digestion and metabolism, and intestinal microbiota in dogs [D].Changchun: Jilin Agricultural University, 2024. | |
| [17] | 仇聪蕊, 舒祥力, 张云飞, 等. 一株猪源罗伊氏黏液乳杆菌的分离鉴定及其益生特性研究 [J]. 中国兽医科学, 2025, 55(9): 1216-1226. |
| Qiu CR, Shu XL, Zhang YF, et al. Isolation, identification and probiotic characteristics analysis of Limosilactobacillus reuteri from porcine [J]. Chin Vet Sci, 2025, 55(9): 1216-1226. | |
| [18] | 林龙镇, 邹卫玲, 李安章, 等. 产酸、耐酸乳酸菌的分离鉴定及益生特性 [J]. 华南农业大学学报, 2018, 39(2): 95-102. |
| Lin LZ, Zou WL, Li AZ, et al. Isolation, identification and probiotic characteristics of acidproducing and acid-resistant Lactobacillus strains [J]. J South China Agric Univ, 2018, 39(2): 95-102. | |
| [19] | Wu JJ, Zhou QY, Liu DM, et al. Evaluation of the safety and probiotic properties of Lactobacillus gasseri LGZ1029 based on whole genome analysis [J]. LWT, 2023, 184: 114759. |
| [20] | Yogeswara IBA, Maneerat S, Haltrich D. Glutamate decarboxylase from lactic acid bacteria-a key enzyme in GABA synthesis [J]. Microorganisms, 2020, 8(12): 1923. |
| [21] | Yang H, He MW, Wu CD. Cross protection of lactic acid bacteria during environmental stresses: Stress responses and underlying mechanisms [J]. LWT, 2021, 144: 111203. |
| [22] | Chintakovid N, Singkhamanan K, Yaikhan T, et al. Probiogenomic analysis of Lactiplantibacillus plantarum SPS109: a potential GABA-producing and cholesterol-lowering probiotic strain [J]. Heliyon, 2024, 10(13): e33823. |
| [23] | Kompramool S, Singkhamanan K, Pomwised R, et al. Genomic insights into Pediococcus pentosaceus ENM104: a probiotic with potential antimicrobial and cholesterol-reducing properties [J]. Antibiotics, 2024, 13(9): 813. |
| [24] | Bäumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut [J]. Nature, 2016, 535(7610): 85-93. |
| [25] | 李彩玉, 刘东慧, 权衡, 等. 益生菌细菌素防御病毒和细菌感染的机制研究 [J]. 中国兽医科学, 2024, 54(6): 808-815. |
| Li CY, Liu DH, Quan H, et al. Mechanism study of probiotics bacteriocins defense against viral and bacterial infections [J]. Chin Vet Sci, 2024, 54(6): 808-815. | |
| [26] | Ginsburg I. Role of lipoteichoic acid in infection and inflammation [J]. Lancet Infect Dis, 2002, 2(3): 171-179. |
| [27] | Averill-bates DA. The antioxidant glutathione [M]//Antioxidants. Amsterdam: Elsevier, 2023: 109-141. |
| [28] | Li XM, Zhang BX, Yan CX, et al. A fast and specific fluorescent probe for thioredoxin reductase that works via disulphide bond cleavage [J]. Nat Commun, 2019, 10: 2745. |
| [29] | Suwannasom N, Kao I, Pruß A, et al. Riboflavin: the health benefits of a forgotten natural vitamin [J]. Int J Mol Sci, 2020, 21(3): 950. |
| [30] | Martin-Gallausiaux C, Marinelli L, Blottière HM, et al. SCFA: mechanisms and functional importance in the gut [J]. Proc Nutr Soc, 2021, 80(1): 37-49. |
| [31] | Smythe P, Efthimiou G. In silico genomic and metabolic atlas of Limosilactobacillus reuteri DSM 20016: an insight into human health [J]. Microorganisms, 2022, 10(7): 1341. |
| [32] | Douillard FP, Ribbera A, Kant R, et al. Comparative genomic and functional analysis of 100 Lactobacillus rhamnosus strains and their comparison with strain GG [J]. PLoS Genet, 2013, 9(8): e1003683. |
| [33] | 苏世友, 滕超, 张维, 等. 细菌Ⅲ型聚酮合酶研究进展 [J]. 中国农业科技导报, 2013, 15(6): 119-129. |
| Su SY, Teng C, Zhang W, et al. Progress on type Ⅲ polyketide synthase from bacteria [J]. J Agric Sci Technol, 2013, 15(6): 119-129. | |
| [34] | Benjdia A, Balty C, Berteau O. Radical SAM enzymes in the biosynthesis of ribosomally synthesized and post-translationally modified peptides (RiPPs) [J]. Front Chem, 2017, 5: 87. |
| [35] | 汪金秀, 张琪, 丁伟, 等. 核糖体肽生物合成中的典型翻译后修饰研究 [J]. 生物技术通报, 2020, 36(10): 215-225. |
| Wang JX, Zhang Q, Ding W, et al. Classic post-translational modification in ribosomally synthesized and post-translationally modified peptides biosynthesis [J]. Biotechnol Bull, 2020, 36(10): 215-225. | |
| [36] | Bonelli RR, Schneider T, Sahl HG, et al. Insights into in vivo activities of lantibiotics from gallidermin and epidermin mode-of-action studies [J]. Antimicrob Agents Chemother, 2006, 50(4): 1449-1457. |
| [37] | 柳少燕, 陈捷胤, 李蕾, 等. 拮抗菌与病原菌碳水化合物酶类比较分析 [J]. 基因组学与应用生物学, 2013, 32(1): 97-104. |
| Liu SY, Chen JY, Li L, et al. Comparative analysis of the carbohydrate-active enzymes between antagonistic microorganism and plant pathogen [J]. Genom Appl Biol, 2013, 32(1): 97-104. | |
| [38] | Oehme DP, Shafee T, Downton MT, et al. Differences in protein structural regions that impact functional specificity in GT2 family β-glucan synthases [J]. PLoS One, 2019, 14(10): e0224442. |
| [39] | Inglin RC, Meile L, Stevens MJA. Clustering of pan- and core-genome of Lactobacillus provides novel evolutionary insights for differentiation [J]. BMC Genom, 2018, 19(1): 284. |
| [1] | LYU Zhen, GAN Tian, HUO Si-yu, ZHAO Chen-di, ZHAO Meng-yao, LI Ya-tao, MA Yu-chao, GENG Yu-qing. Identification of Surfactin-producing Bacillus Velezensis C5A-1 and Evaluation of the Plant Growth-promoting Effects of Its Surfactin [J]. Biotechnology Bulletin, 2025, 41(9): 265-276. |
| [2] | SU Xiu-min, HAN Wen-qing, WANG Jiao, LI Peng, WANG Qiu-lan, LI Wan-xing, CAO Jin-jun. Isolation, Identification, Biological Characteristics and Biocontrol Effects of Trichoderma harzianum M408 against Tomato Early Blight [J]. Biotechnology Bulletin, 2025, 41(9): 277-288. |
| [3] | LIAN Shao-jie, TANG Sheng-shuo, KANG Chuan-li, LIU Lei, ZHENG De-qiang, DU Shuai, TANG Li-wei, ZHANG Mei-xia, LIU Qiang. Isolation, Identification, Optimization of Fermentation Conditions of High-yield Tremella fuciformis Polysaccharides Enzyme-producing Strain and Its Enzyme Characteristics Analysis [J]. Biotechnology Bulletin, 2025, 41(9): 302-313. |
| [4] | ZHANG Ru, LI Yi-ming, ZHANG Tong-xi, SUN Zhan-bin, REN Qing, PAN Han-xu. Isolation and Identification of a High-yielding Magnolol and Honokiol Strain from Magnolia officinalis and Optimization of the “Sweating” Process [J]. Biotechnology Bulletin, 2025, 41(8): 322-334. |
| [5] | XIANG Bo-ka, ZHOU Zuan-zuan, FENG Jia-hui, XIA Chen, LI Qi, CHEN Chun. Isolation and Identification of a Fungus from Moldy Tobacco Leaf and Study on Its Mold-causing Factors [J]. Biotechnology Bulletin, 2025, 41(2): 321-330. |
| [6] | XU Yuan-zhi, HU Shan, DAI Si-ze, YOU Shuai, ZHENG Ming-ming, SHAN Kai. Discovery and Evaluation of High-activity Lipase and Its Application in Diacylglycerol Synthesis [J]. Biotechnology Bulletin, 2025, 41(11): 177-189. |
| [7] | MIAO Hao-xiang, ZHANG Ying, GUO Shi-peng, ZHANG Jian. Whole Genome Sequencing and Comparative Genomic Analysis of a High-yielding γ-aminobutyric Acid-producing Lactobacillus brevis TCCC13007 [J]. Biotechnology Bulletin, 2025, 41(11): 166-176. |
| [8] | ZHANG Ting, WAN Yu-xin, XU Wei-hui, WANG Zhi-gang, CHEN Wen-jing, HU Yun-long. Growth-promoting Effects of a Rhizosphere Growth-promoting Bacterium Leclercia adecarboxylata LN01 in Maize Plants and Its Whole-genome Analysis [J]. Biotechnology Bulletin, 2025, 41(1): 263-275. |
| [9] | ZHANG Ya-ya, LI Pan-pan, GAO Hui-hui, JIA Chen-bo, XU Chun-yan. Exploring on the Pathogenesis of Root Rot of Lycium barbarum cv. ‘Ningqi-5' Based on the Rhizoplane Fungal Community and Pathogens Identification [J]. Biotechnology Bulletin, 2024, 40(9): 238-248. |
| [10] | WANG Fang, YU Lu, QI Ze-zheng, ZHOU Chang-jun, YU Ji-dong. Screening and Biocontrol Effect of Antagonistic Bacteria against Soybean Root Rot [J]. Biotechnology Bulletin, 2024, 40(7): 216-225. |
| [11] | SUN Ya-nan, WANG Chun-xue, WANG Xin, DU Bing-hai, LIU Kai, WANG Cheng-qiang. Biocontrol Characteristics of Bacillus atrophaeus CNY01 and Its Salt-resistant and Growth-promoting Effect on Maize Seedling [J]. Biotechnology Bulletin, 2024, 40(5): 248-260. |
| [12] | WANG Lu, LIU Meng-yu, ZHANG Fu-yuan, JI Shou-kun, WANG Yun, ZHANG Ying-jie, DUAN Chun-hui, LIU Yue-qin, YAN Hui. Isolation and Identification of Rumen Skatole-degrading Bacteria and Analysis on Their Degradation Characteristics [J]. Biotechnology Bulletin, 2024, 40(3): 305-311. |
| [13] | RAO Zi-huan, XIE Zhi-xiong. Isolation and Identification of a Cellulose-degrading Strain of Olivibacter jilunii and Analysis of Its Degradability [J]. Biotechnology Bulletin, 2023, 39(8): 283-290. |
| [14] | YOU Zi-juan, CHEN Han-lin, DENG Fu-cai. Research Progress in the Extraction and Functional Activities of Bioactive Peptides from Fish Skin [J]. Biotechnology Bulletin, 2023, 39(7): 91-104. |
| [15] | CHE Yong-mei, GUO Yan-ping, LIU Guang-chao, YE Qing, LI Ya-hua, ZHAO Fang-gui, LIU Xin. Isolation and Identification of Bacterial Strain C8 and B4 and Their Halotolerant Growth-promoting Effects and Mechanisms [J]. Biotechnology Bulletin, 2023, 39(5): 276-285. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||