








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (7): 146-155.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0362
Previous Articles Next Articles
SHAN Cao-mei( ), YE Lei, ZHANG Lian-hu, KUANG Wei-gang, SUN Xiao-tang, MA Jian, CUI Ru-qiang(
), YE Lei, ZHANG Lian-hu, KUANG Wei-gang, SUN Xiao-tang, MA Jian, CUI Ru-qiang( )
)
Received:2021-03-22
Online:2021-07-26
Published:2021-08-13
Contact:
CUI Ru-qiang
E-mail:StrawM@126.com;cuiruqiang@jxau.edu.cn
SHAN Cao-mei, YE Lei, ZHANG Lian-hu, KUANG Wei-gang, SUN Xiao-tang, MA Jian, CUI Ru-qiang. Cloning,and Functional Analysis of Gene OsRAI1 Resistant to Hirschmanniella mucronate in Rice[J]. Biotechnology Bulletin, 2021, 37(7): 146-155.
| 名称Name | 序列 Sequences(5'-3') | 作用 Function |
|---|---|---|
| OsRAI1-F | ATGGAGCTTGACGAG | 基因克隆 Gene cloning |
| OsRAI1-R | CAGACACCTTCCGCC | |
| T7 | CCTATAGTGAGTCGTATTA | 原核表达载体鉴定 Identification of prokaryotic expression vectors |
| T7ter | CTAGCATAACCCCTTGGGGC | |
| pET28a-OsRAI1-F | CAGCAAATGGGTCGCGGATCCATGGAGCTTGACGAG | 添加酶切位点 Adding enzyme cutting site |
| pET28a-OsRAI1-R | ACTCGAGCACCACCAGCGGCCGCCAGACACCTTCCGCC | |
| 1302-OsRAI1-F | ACCATGGTAGATCTGACTAGTATGGAGCTTGACGAGGAGTCC | 添加酶切位点 Adding enzyme cutting site |
| 1302-OsRAI1-R | AAGTTCTTCTCCTTTACTAGTCAGACACCTTCCGCCATAGC | |
| pCA-F | TGAGACTTTTCAACAAAGGGTAATATCCGG | 过表达载体鉴定 Identification of overexpression vectors |
| pCA-R | GATCTAGTAACATAGATGACACCGC |
Table 1 Information of PCR primers
| 名称Name | 序列 Sequences(5'-3') | 作用 Function |
|---|---|---|
| OsRAI1-F | ATGGAGCTTGACGAG | 基因克隆 Gene cloning |
| OsRAI1-R | CAGACACCTTCCGCC | |
| T7 | CCTATAGTGAGTCGTATTA | 原核表达载体鉴定 Identification of prokaryotic expression vectors |
| T7ter | CTAGCATAACCCCTTGGGGC | |
| pET28a-OsRAI1-F | CAGCAAATGGGTCGCGGATCCATGGAGCTTGACGAG | 添加酶切位点 Adding enzyme cutting site |
| pET28a-OsRAI1-R | ACTCGAGCACCACCAGCGGCCGCCAGACACCTTCCGCC | |
| 1302-OsRAI1-F | ACCATGGTAGATCTGACTAGTATGGAGCTTGACGAGGAGTCC | 添加酶切位点 Adding enzyme cutting site |
| 1302-OsRAI1-R | AAGTTCTTCTCCTTTACTAGTCAGACACCTTCCGCCATAGC | |
| pCA-F | TGAGACTTTTCAACAAAGGGTAATATCCGG | 过表达载体鉴定 Identification of overexpression vectors |
| pCA-R | GATCTAGTAACATAGATGACACCGC |
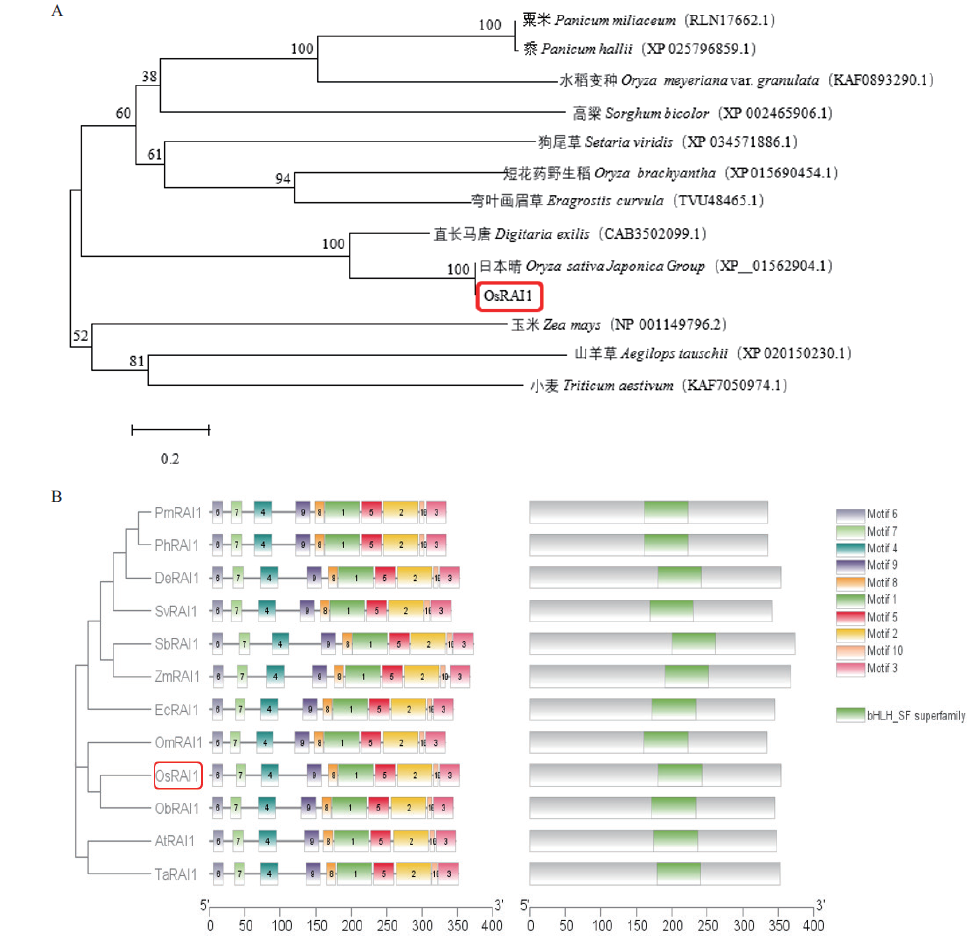
Fig. 5 Development and evolution of OsRAI1 protein system A: Phylogenetic tree of OsRAI1. B: Conserved motif element prediction of OsRAI1 protein sequences
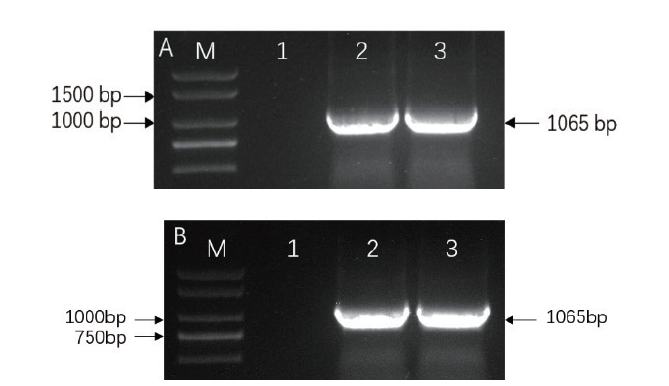
Fig. 6 Electropherogram of PCR product for OsRAI1 gene A:Amplified product of gene OsRAI1 via prokaryotic expression; B:Amplified product of gene OsRAI1 via overexpression. M:2 000 marker; 1-3:Amplified product
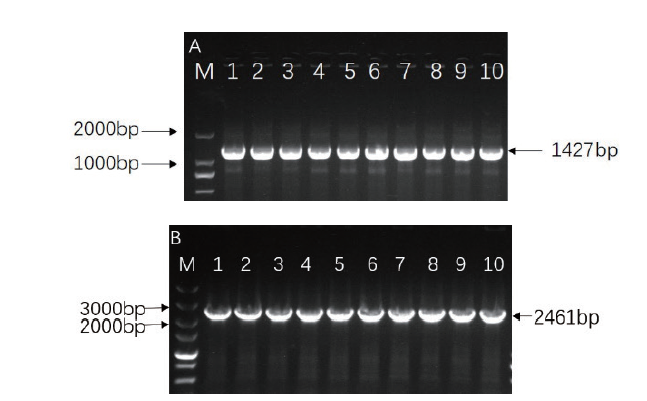
Fig. 7 Identification of positive clones of OsRAI1 gene via vectors A:Identification of positive clones of gene OsRAI1 via prokaryotic expression vector; M:2000 marker. 1-10:PCR product of single colony. B:Identification of positive clones of gene OsRAI1 via overexpression vector; M:5000 marker. 1-10:PCR product of single colony

Fig. 8 Subcellular localization of gene OsRAI1 in the leaves of Nicotiana benthamiana A:Target protein fluorescence channel. B:Chloroplast fluorescence channel. C:Brightfield. D:Overlay chart

Fig. 9 WB analysis of recombinant protein expression products induced by different concentrations of IPTG M:Protein marker. 1:Whole bacteria induced by 1 mmol/L. 2:Supernatant induced by 0.8 mmol/L. 3:Whole bacteria induced by 0.8 mmol/L. 4:Supernatant induced by 0.6 mmol/L. 5:Whole bacteria induced by 0.6 mmol/L. 6:Supernatant induced by 0.2 mmol/L. 7:Whole bacteria induced by 0.2 mmol/L

Fig. 10 SDS-PAGE analysis of recombinant protein expression products induced by different inducing time and temperature M:Protein Marker. 1:Whole bacteria induced by 37℃ for 3 h. 2:Whole bacteria induced by 37℃ for 6 h. 3:Whole bacteria induced by 37℃ for 18 h. 4:Whole bacteria induced by 28℃ for 3 h. 5:Whole bacteria induced by 28℃ for 6 h. 6:Whole bacteria induced by 28℃ for 18 h. 7:Whole bacteria induced by 16℃ for 3 h. 8:Whole bacteria induced by 16℃ for 6 h. 9:Whole bacteria induced by 16℃ for 18 h. 10:Supernatant induced by 37℃ for 3 h. 11:Supernatant induced by 37℃ for 6 h. 12:Supernatant induced by 37℃ for 18 h. 13:Supernatant induced by 28℃ for 3 h. 14:Supernatant induced by 28℃ for 6 h. 15:Supernatant induced by 28℃ for 18 h. 16:Supernatant induced by 16℃ for 3 h. 17:Supernatant induced by 16℃ for 6 h. 18:Supernatant induced by 16℃ for 18 h
| 基因名称 Gene name | 蛋白描述 Protein description | 功能结构域 Functional domain | 氨基酸数/个 Number of amino acids | 互作系数 Interaction coefficient |
|---|---|---|---|---|
| Os05T0586300-00 | Expressed protein;Os03g0137400 protein | HLH | 387 | 0.644 |
| Os04T0486400-01 | OJ000223_09.13 protein;Os04g0486400 protein | SANT | 385 | 0.612 |
| Os03T0137400-01 | Expressed protein;Os03g0137400 protein | transmembrane helix region | 792 | 0.578 |
| Os01T0752500-00 | Putative ethylene responsive protein Os01g0752500 protein | AP2 | 212 | 0.570 |
| Os06T0164400-01 | annotation not available;Os06g0164400 protein | coiled coil region;HLH | 197 | 0.503 |
Table 2 Functional interaction protein prediction of OsRAI1
| 基因名称 Gene name | 蛋白描述 Protein description | 功能结构域 Functional domain | 氨基酸数/个 Number of amino acids | 互作系数 Interaction coefficient |
|---|---|---|---|---|
| Os05T0586300-00 | Expressed protein;Os03g0137400 protein | HLH | 387 | 0.644 |
| Os04T0486400-01 | OJ000223_09.13 protein;Os04g0486400 protein | SANT | 385 | 0.612 |
| Os03T0137400-01 | Expressed protein;Os03g0137400 protein | transmembrane helix region | 792 | 0.578 |
| Os01T0752500-00 | Putative ethylene responsive protein Os01g0752500 protein | AP2 | 212 | 0.570 |
| Os06T0164400-01 | annotation not available;Os06g0164400 protein | coiled coil region;HLH | 197 | 0.503 |
| [1] |
de Silva WSI, Perera MMN, Perera KLNS, et al. In silico analysis of osr40c1 promoter sequence isolated from indica variety pokkali[J]. Rice Sci, 2017, 24(4):228-234.
doi: 10.1016/j.rsci.2016.11.002 URL |
| [2] |
Wani SH, Kumar Sah S. Biotechnology and abiotic stress tolerance in rice[J]. J Rice Res, 2014, 2(2):e105. DOI: 10.4172/jrr.1000e105.
doi: 10.4172/jrr.1000e105 |
| [3] | 曾红根. 江西省高产优质直播水稻栽培技术及病虫草害防治探析[J]. 种子科技, 2020, 38(20):98-99. |
| Zeng HG. Analysis on cultivation technology of high yield and high quality direct seeding rice and prevention and control of pests and weeds in jiangxi province[J]. Seed Science & Technology, 2020, 38(20):98-99. | |
| [4] | 汪家旭, 潘沧桑. 潜根线虫的种类[J]. 厦门大学学报:自然科学版, 1999, 38(2):145-152. |
| Wang JX, Pan CS. Studies of rice root Nematodes(Hirschmanniella species)[J]. J Xiamen Univ:Nat Sci, 1999, 38(2):145-152. | |
| [5] |
Elling AA. Major emerging problems with minor Meloidogyne species[J]. Phytopathology, 2013, 103(11):1092-1102.
doi: 10.1094/PHYTO-01-13-0019-RVW pmid: 23777404 |
| [6] | 张绍升, 李茂胜, 严叔平. 水稻潜根线虫的致病性和综合防治技术[J]. 中国水稻科学, 1998, 12(1):31-34. |
| Zhang SS, Li MS, Yan SP. The pathogenicity and integrated control measures of rice root Nematodes[J]. Chin J Rice Sci, 1998, 12(1):31-34. | |
| [7] |
Sun XT, Zhang L, Tang ZQ, et al. Transcriptome analysis of roots from resistant and susceptible rice varieties infected with Hirschmanniella mucronata[J]. FEBS Open Bio, 2019, 9(11):1968-1982.
doi: 10.1002/feb4.v9.11 URL |
| [8] | 于冰, 田烨, 李海英, 等. 植物bHLH转录因子的研究进展[J]. 中国农学通报, 2019, 35(9):75-80. |
| Yu B, Tian Y, Li HY, et al. Research progress of plant bHLH transc-ription factor[J]. Chin Agric Sci Bull, 2019, 35(9):75-80. | |
| [9] |
Li X, Duan X, Jiang H, et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis[J]. Plant Physiol, 2006, 141(4):1167-1184.
doi: 10.1104/pp.106.080580 URL |
| [10] | 刘晓月, 王文生, 傅彬英. 植物bHLH转录因子家族的功能研究进展[J]. 生物技术进展, 2011, 1(6):391-397. |
| Liu XY, Wang WS, Fu BY. Research progress of plant bHLH transcription factor family[J]. Curr Biotechnol, 2011, 1(6):391-397. | |
| [11] |
Thorstensen T, Grini PE, Mercy IS, et al. The Arabidopsis SET-domain protein ASHR3 is involved in stamen development and interacts with the bHLH transcription factor ABORTED MICROSPORES(AMS)[J]. Plant Mol Biol, 2008, 66(1/2):47-59.
doi: 10.1007/s11103-007-9251-y URL |
| [12] | 王艳敏, 白卉, 曹焱. bHLH转录因子研究进展及其在植物抗逆中的应用[J]. 安徽农业科学, 2015, 43(21):34-35, 50. |
| Wang YM, Bai H, Cao Y. Research progress of bHLH transcription factor and application in plant abiotic stress tolerance[J]. J Anhui Agric Sci, 2015, 43(21):34-35, 50. | |
| [13] |
Wang YJ, Zhang ZG, He XJ, et al. A rice transcription factor OsbHLH1 is involved in cold stress response[J]. Theor Appl Genet, 2003, 107(8):1402-1409.
doi: 10.1007/s00122-003-1378-x URL |
| [14] |
Ortolan F, Fonini LS, Pastori T, et al. Tightly controlled expression of OsbHLH35 is critical for anther development in rice[J]. Plant Sci, 2021, 302:110716.
doi: 10.1016/j.plantsci.2020.110716 URL |
| [15] |
Wang L, Ying Y, Narsai R, et al. Identification of OsbHLH133 as a regulator of iron distribution between roots and shoots in Oryza sativa[J]. Plant Cell Environ, 2013, 36(1):224-236.
doi: 10.1111/pce.2013.36.issue-1 URL |
| [16] |
Lee J, Moon S, Jang S, et al. OsbHLH073 negatively regulates internode elongation and plant height by modulating GA homeostasis in rice[J]. Plants, 2020, 9(4):547.
doi: 10.3390/plants9040547 URL |
| [17] | 邵雅芳, 徐非非, 唐富福, 等. 水稻花青素合成相关基因的时空表达研究[J]. 核农学报, 2013, 27(1):9-14. |
| Shao YF, Xu FF, Tang FF, et al. The temporal and spatial expression pattern of anthocyanin related genes in RICE(Oryza sativa L.)[J]. J Nucl Agric Sci, 2013, 27(1):9-14. | |
| [18] |
Carretero-Paulet L, Galstyan A, Roig-Villanova I, et al. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae[J]. Plant Physiol, 2010, 153(3):1398-1412.
doi: 10.1104/pp.110.153593 pmid: 20472752 |
| [19] |
Chen ZH, Nimmo GA, et al. BHLH32 modulates several biochemical and morphological processes that respond to Pi starvation in Arabidopsis[J]. Biochem J, 2007, 405(1):191-198.
doi: 10.1042/BJ20070102 URL |
| [20] |
Sun XH, Copeland NG, Jenkins NA, et al. Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins[J]. Mol Cell Biol, 1991, 11(11):5603-5611.
pmid: 1922066 |
| [21] | Qin Y, L i X, Guo M, et al. Regulation of salt and ABA responses by CIPK14, a calcium sensor interacting protein kinase in Arabidopsis[J]. Sci China C Life Sci, 2008, 51(5):391-401. |
| [1] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [2] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [3] | MEI Huan, LI Yue, LIU Ke-meng, LIU Ji-hua. Study on the Biosynthesis of l-SLR by Efficient Prokaryotic Expression of Berberine Bridge Enzyme [J]. Biotechnology Bulletin, 2023, 39(7): 277-287. |
| [4] | YANG Xu-yan, ZHAO Shuang, MA Tian-yi, BAI Yu, WANG Yu-shu. Cloning of Three Cabbage WRKY Genes and Their Expressions in Response to Abiotic Stress [J]. Biotechnology Bulletin, 2023, 39(11): 261-269. |
| [5] | GUO Zhi-hao, JIN Ze-xin, LIU Qi, GAO Li. Bioinformatics Analysis, Subcellular Localization and Toxicity Verification of Effector g11335 in Tilletia contraversa Kühn [J]. Biotechnology Bulletin, 2022, 38(8): 110-117. |
| [6] | SUO Qing-qing, WU Nan, YANG Hui, LI Li, WANG Xi-feng. Prokaryotic Expression,Antibody Preparation and Application of Rice Caffeoyl Coenzyme A-O-methyltransferase Gene [J]. Biotechnology Bulletin, 2022, 38(8): 135-141. |
| [7] | QIN Xue-jing, WANG Yu-han, CAO Yi-bo, ZHANG Ling-yun. Prokaryotic Expression and Preparation of Polyclonal Antibody of PwHAP5 Gene in Picea wilsonii [J]. Biotechnology Bulletin, 2022, 38(8): 142-149. |
| [8] | WANG Guang-li, FAN Chan, WANG Hui, LU Hui-fang, XIA Ling-yin, HUANG Jian, MIN Xun. Prokaryotic Expression,Purification,Identification,and Polyclonal Antibody Preparation of Vibrio cholerae Hemolysin HlyA [J]. Biotechnology Bulletin, 2022, 38(7): 269-277. |
| [9] | YANG Jia-bao, ZHOU Zhi-ming, ZHANG Zhan, FENG Li, SUN Li. Cloning,Expression of Helianthus annuus HaLACS1 Gene and Identification of Its Functional Complementation in Saccharomyces cerevisiae [J]. Biotechnology Bulletin, 2022, 38(6): 147-156. |
| [10] | HAO Qing-qing, YAO Sheng, LIU Jia-he, CHEN Pei-zhen, ZHANG Meng-yang, JI Kong-shu. Cloning and Expression Analysis of NAC Transcription Factor PmNAC8 in Pinus massoniana [J]. Biotechnology Bulletin, 2022, 38(4): 202-216. |
| [11] | WANG Qiao-ju, HU Yu-meng, WEN Ya-ya, SONG Li, MENG Chuang, PAN Zhi-ming, JIAO Xin-an. Expression and Activity Identification of SARS-CoV-2 S1 Protein [J]. Biotechnology Bulletin, 2022, 38(3): 157-163. |
| [12] | FENG Jian-ying, LI Li-qin, LU Li-ming. Genome-wide Identification and Expression Analysis of the bHLH Transcription Factor Family in Solanum tuberosum [J]. Biotechnology Bulletin, 2022, 38(2): 21-33. |
| [13] | DANG Yuan, LI Wei, MIAO Xiang, XIU Yu, LIN Shan-zhi. Cloning of Oleosin Gene PsOLE4 from Prunus sibirica and Its Regulatory Function Analysis for Oil Accumulation [J]. Biotechnology Bulletin, 2022, 38(11): 151-161. |
| [14] | LUO Ying, TANG Zhi, WANG Fan, LIU Xiao-xia, LUO Xiao-fang, HE Fu-lin. Cloning and Response Analysis to Abiotic Stress of GbR2R3-MYB1 Gene from Ginkgo biloba [J]. Biotechnology Bulletin, 2022, 38(10): 184-194. |
| [15] | SHEN Jun-qiang, ZHANG Li-ping, YU Rui-ming, WANG Yong-lu, PAN Li, LIU Xia, LIU Xin-sheng. Porcine Kobuvirus Structural Proteins VP0 and VP1 Prokaryotic Expression and Establishment of Indirect ELISA Method [J]. Biotechnology Bulletin, 2022, 38(10): 243-253. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||