








Biotechnology Bulletin ›› 2021, Vol. 37 ›› Issue (8): 141-151.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0068
Previous Articles Next Articles
MA Ya-nan1( ), LU Xu2, WEI Yun-chun1, LI Kang1, WEI Ruo-nan1, LI Sheng1(
), LU Xu2, WEI Yun-chun1, LI Kang1, WEI Ruo-nan1, LI Sheng1( ), MA Shao-ying3(
), MA Shao-ying3( )
)
Received:2021-01-17
Online:2021-08-26
Published:2021-09-10
Contact:
LI Sheng,MA Shao-ying
E-mail:m568518983@qq.com;lish@gsau.edu.cn;mashy@gsau.edu.cn
MA Ya-nan, LU Xu, WEI Yun-chun, LI Kang, WEI Ruo-nan, LI Sheng, MA Shao-ying. Identification and Tissue Specific Expression Analysis of AKR Gene Family in Grape[J]. Biotechnology Bulletin, 2021, 37(8): 141-151.
| 基因Gene | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| VvAKR01 | AGGAGGCAGCAGTGTCAAAATGG | CCAGCCACCGCATCATCATCTC |
| VvAKR02 | ACCATTGAAAGCCTCAGCCAGTG | GGCGTTGTCAGAGGGTTATCGTC |
| VvAKR03 | TTGTGCTGAGGAAGCTGGGTAAAC | GCCGCAGAGTGGGAACAAGAAG |
| VvAKR04 | AGCGGTGATTTGAATGCCTTTTGC | GGCGGTCAATACGGTGGAGATG |
| VvAKR05 | AGCGGGTTCATCTCCACTTGATTG | GGAAGGCATGGAAGAGTGTCAGAG |
| VvAKR06 | TCTGCTCTGTCTTGTGCGAAACC | GAACCAACCCCATACCCTAAAGCC |
| VvAKR07 | TGCTGCCAAAGTGGGTTCATCTC | GTCAGAAGCTCGGTCTCACCAAA |
| VvAKR08 | AAAGCTTTGGTGCACCGACG | TCCCAGGCTTCAAGCTCACC |
| VvAKR09 | TAGCCGCCTTAAGAGCCTGTCC | ACGCTGTCTTCACCGGCAATCG |
| VvAKR10 | GACACTGTGACCGCTCTGAATACC | CTCTCGGCGTACTCTCCTCTTGG |
| VvAKR11 | CTTGGATGACAGCAGGAGGAACAC | AGCACATGGAGAGCAATGGAAGC |
| VvAKR12 | AGTGCTTCCATTGCTCTCCATGTG | CTTATCCATTGGCCGGTCCGTATG |
| VvAKR13 | CTGCCTGCTCTTGCCAGATGTG | CCGTGTTCCTCCTGCTGTCAATC |
| UBI | GCTCGCTGTTTTTGCAGTTCTAC | AACATAGGTGAGGCCGCACTT |
Table 1 Real-time fluorescence quantitative primers of AKR gene family in grape
| 基因Gene | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| VvAKR01 | AGGAGGCAGCAGTGTCAAAATGG | CCAGCCACCGCATCATCATCTC |
| VvAKR02 | ACCATTGAAAGCCTCAGCCAGTG | GGCGTTGTCAGAGGGTTATCGTC |
| VvAKR03 | TTGTGCTGAGGAAGCTGGGTAAAC | GCCGCAGAGTGGGAACAAGAAG |
| VvAKR04 | AGCGGTGATTTGAATGCCTTTTGC | GGCGGTCAATACGGTGGAGATG |
| VvAKR05 | AGCGGGTTCATCTCCACTTGATTG | GGAAGGCATGGAAGAGTGTCAGAG |
| VvAKR06 | TCTGCTCTGTCTTGTGCGAAACC | GAACCAACCCCATACCCTAAAGCC |
| VvAKR07 | TGCTGCCAAAGTGGGTTCATCTC | GTCAGAAGCTCGGTCTCACCAAA |
| VvAKR08 | AAAGCTTTGGTGCACCGACG | TCCCAGGCTTCAAGCTCACC |
| VvAKR09 | TAGCCGCCTTAAGAGCCTGTCC | ACGCTGTCTTCACCGGCAATCG |
| VvAKR10 | GACACTGTGACCGCTCTGAATACC | CTCTCGGCGTACTCTCCTCTTGG |
| VvAKR11 | CTTGGATGACAGCAGGAGGAACAC | AGCACATGGAGAGCAATGGAAGC |
| VvAKR12 | AGTGCTTCCATTGCTCTCCATGTG | CTTATCCATTGGCCGGTCCGTATG |
| VvAKR13 | CTGCCTGCTCTTGCCAGATGTG | CCGTGTTCCTCCTGCTGTCAATC |
| UBI | GCTCGCTGTTTTTGCAGTTCTAC | AACATAGGTGAGGCCGCACTT |
| 基因Gene | 登录号 Gene accession No. | 染色体定位 Chromosomal location | 全长Full length/bp | CDS/bp | 氨基酸 Amino acid | 分子量 Molecular weight/bp | 等电点 pI | 不稳定系数 Instability index | 脂肪系数Aliphatic index | 平均亲水性Hydropathicity |
|---|---|---|---|---|---|---|---|---|---|---|
| VvAKR01 | GSVIVT01009396001 | chr18:7983438..7986889 | 3452 | 969 | 322 | 36192.49 | 5.9 | 38.98 | 89.66 | -0.238 |
| VvAKR02 | GSVIVT01010238001 | chr1:18082562..18106134 | 23573 | 1002 | 333 | 36674.09 | 7.07 | 34.21 | 90.75 | -0.213 |
| VvAKR03 | GSVIVT01011576001 | chr1:6306239..6307845 | 1607 | 867 | 288 | 32083.49 | 9.01 | 42.94 | 96.88 | -0.086 |
| VvAKR04 | GSVIVT01011577001 | chr1:6298552..6305704 | 7153 | 912 | 303 | 34017.51 | 6.54 | 47.47 | 89.8 | -0.164 |
| VvAKR05 | GSVIVT01011579001 | chr1:6289243..6290820 | 1578 | 951 | 316 | 35455.01 | 5.85 | 51.55 | 95.73 | -0.206 |
| VvAKR06 | GSVIVT01011582001 | chr1:6274423..6285766 | 11344 | 1293 | 430 | 48655.51 | 7.1 | 43.48 | 94.12 | -0.184 |
| VvAKR07 | GSVIVT01011583001 | chr1:6264420..6272529 | 8110 | 909 | 302 | 34322.71 | 6.34 | 27.01 | 92.02 | -0.274 |
| VvAKR08 | GSVIVT01011585001 | chr1:6241008..6248273 | 7266 | 1092 | 363 | 40730.95 | 5.98 | 49.92 | 95.92 | -0.228 |
| VvAKR09 | GSVIVT01030263001 | chr8:9745143..9747339 | 2197 | 969 | 322 | 35975.24 | 6.17 | 45.07 | 83.57 | -0.319 |
| VvAKR10 | GSVIVT01034098001 | chr8:15157629..15161115 | 3487 | 948 | 315 | 35068.99 | 5.83 | 50.57 | 89.46 | -0.243 |
| VvAKR11 | GSVIVT01034099001 | chr8:15153012..15157096 | 4085 | 828 | 275 | 30440 | 6.45 | 46.14 | 87.56 | -0.204 |
| VvAKR12 | GSVIVT01034101001 | chr8:15144714..15149849 | 5136 | 903 | 300 | 33663.65 | 6.76 | 42.71 | 88.37 | -0.317 |
Table 2 Physical and chemical property of AKR genes in grape
| 基因Gene | 登录号 Gene accession No. | 染色体定位 Chromosomal location | 全长Full length/bp | CDS/bp | 氨基酸 Amino acid | 分子量 Molecular weight/bp | 等电点 pI | 不稳定系数 Instability index | 脂肪系数Aliphatic index | 平均亲水性Hydropathicity |
|---|---|---|---|---|---|---|---|---|---|---|
| VvAKR01 | GSVIVT01009396001 | chr18:7983438..7986889 | 3452 | 969 | 322 | 36192.49 | 5.9 | 38.98 | 89.66 | -0.238 |
| VvAKR02 | GSVIVT01010238001 | chr1:18082562..18106134 | 23573 | 1002 | 333 | 36674.09 | 7.07 | 34.21 | 90.75 | -0.213 |
| VvAKR03 | GSVIVT01011576001 | chr1:6306239..6307845 | 1607 | 867 | 288 | 32083.49 | 9.01 | 42.94 | 96.88 | -0.086 |
| VvAKR04 | GSVIVT01011577001 | chr1:6298552..6305704 | 7153 | 912 | 303 | 34017.51 | 6.54 | 47.47 | 89.8 | -0.164 |
| VvAKR05 | GSVIVT01011579001 | chr1:6289243..6290820 | 1578 | 951 | 316 | 35455.01 | 5.85 | 51.55 | 95.73 | -0.206 |
| VvAKR06 | GSVIVT01011582001 | chr1:6274423..6285766 | 11344 | 1293 | 430 | 48655.51 | 7.1 | 43.48 | 94.12 | -0.184 |
| VvAKR07 | GSVIVT01011583001 | chr1:6264420..6272529 | 8110 | 909 | 302 | 34322.71 | 6.34 | 27.01 | 92.02 | -0.274 |
| VvAKR08 | GSVIVT01011585001 | chr1:6241008..6248273 | 7266 | 1092 | 363 | 40730.95 | 5.98 | 49.92 | 95.92 | -0.228 |
| VvAKR09 | GSVIVT01030263001 | chr8:9745143..9747339 | 2197 | 969 | 322 | 35975.24 | 6.17 | 45.07 | 83.57 | -0.319 |
| VvAKR10 | GSVIVT01034098001 | chr8:15157629..15161115 | 3487 | 948 | 315 | 35068.99 | 5.83 | 50.57 | 89.46 | -0.243 |
| VvAKR11 | GSVIVT01034099001 | chr8:15153012..15157096 | 4085 | 828 | 275 | 30440 | 6.45 | 46.14 | 87.56 | -0.204 |
| VvAKR12 | GSVIVT01034101001 | chr8:15144714..15149849 | 5136 | 903 | 300 | 33663.65 | 6.76 | 42.71 | 88.37 | -0.317 |
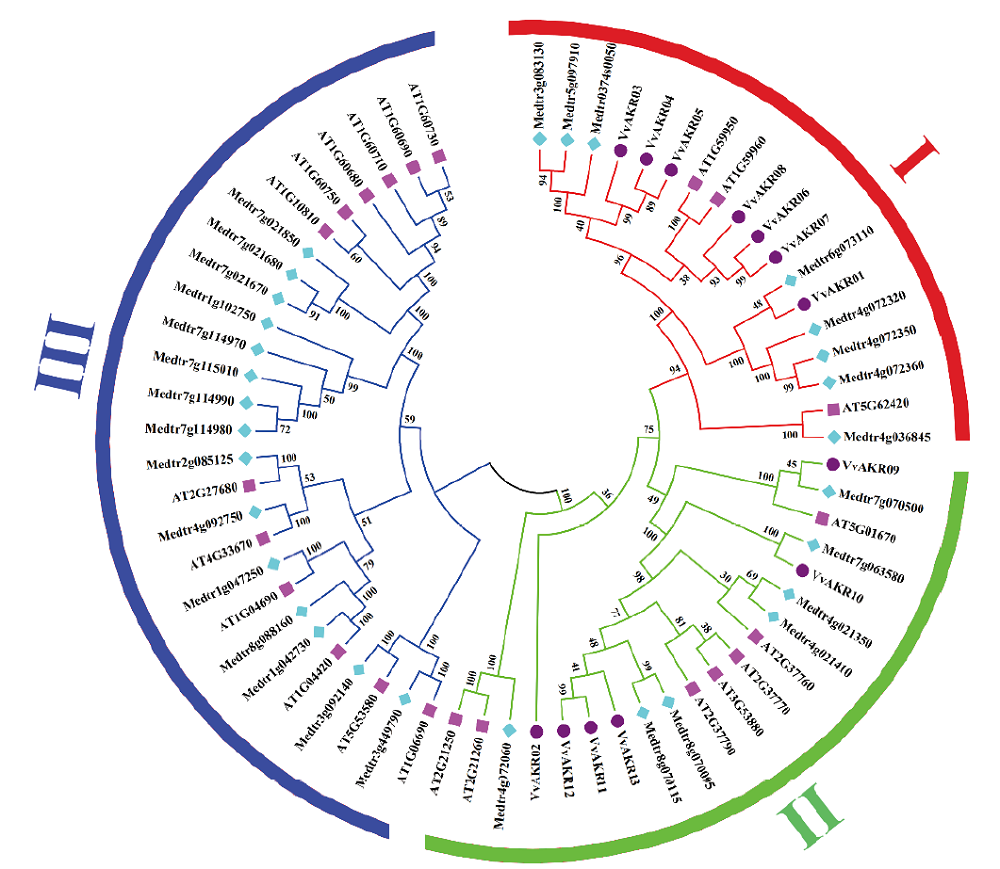
Fig.1 Phylogenetic tree of AKR genes in Arabidopsis,alfalfa and grape Pink squares represent Arabidopsis, light blue diamonds represent alfalfa, and purple circles refer to grapes
| 基因 Gene | 分子式 Formula | α-螺旋 Alpha helix/% | β-转角 Beta turn/% | 扩展链结构 Hydropathicity/% | 不规则卷曲 Random coil/% | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|
| VvAKR01 | C1621H2533N437O473S15 | 42.55 | 4.04 | 14.29 | 39.13 | Nucleus,cytosol,cytoskeleton and chloroplast |
| VvAKR02 | C1630H2584N452O480S15 | 37.84 | 6.01 | 15.32 | 40.84 | Nucleus,cytosol,cytoskeleton and chloroplast |
| VvAKR03 | C1437H2314N388O409S16 | 41.32 | 4.51 | 18.06 | 36.11 | Nucleus,membrane,cytosol,chloroplast and mitochondrion,peroxisome |
| VvAKR04 | C1533H2418N402O437S17 | 42.57 | 4.62 | 13.86 | 38.94 | Nucleus,cytosol,cytoskeleton and chloroplast |
| VvAKR05 | C1584H2534N424O466S15 | 43.04 | 3.80 | 15.51 | 37.66 | Extracellular,cytosol,chloroplast,mitochondrion and endoplasmic reticulum |
| VvAKR06 | C2207H3481N571O627S19 | 34.42 | 3.95 | 17.44 | 44.19 | Nucleus,membrane,cytosol,cytoskeleton,chloroplast and mitochondrion |
| VvAKR07 | C1560H2445N401O445S12 | 41.39 | 5.30 | 13.58 | 39.74 | Nucleus,extracellular,cytosol and cytoskeleton |
| VvAKR08 | C1829H2900N490O533S14 | 42.15 | 4.41 | 13.50 | 39.94 | Nucleus,membrane,cytosol and cytoskeleton |
| VvAKR09 | C1601H2518N442O469S16 | 40.37 | 8.39 | 14.60 | 36.65 | Nucleus,membrane,cytosol,cytoskeleton and chloroplast |
| VvAKR10 | C1582H2451N419O463S10 | 42.54 | 6.67 | 13.97 | 36.83 | Nucleus,cytosol and chloroplast |
| VvAKR11 | C1378H2147N363O395S10 | 43.27 | 5.09 | 14.18 | 37.45 | Nucleus,cytosol,cytoskeleton,endoplasmic reticulum and vacuole |
| VvAKR12 | C1518H2375N411O435S10 | 43.00 | 6.67 | 12.00 | 38.33 | Nucleus,membrane,cytosol,cytoskeleton and vacuole |
| VvAKR013 | C1872H2916N492O531S11 | 48.91 | 6.25 | 11.41 | 33.42 | Nucleus,cytosol and chloroplast |
Table 3 Secondary structure and Subcellular location prediction of AKR genes in grape
| 基因 Gene | 分子式 Formula | α-螺旋 Alpha helix/% | β-转角 Beta turn/% | 扩展链结构 Hydropathicity/% | 不规则卷曲 Random coil/% | 亚细胞定位 Subcellular localization |
|---|---|---|---|---|---|---|
| VvAKR01 | C1621H2533N437O473S15 | 42.55 | 4.04 | 14.29 | 39.13 | Nucleus,cytosol,cytoskeleton and chloroplast |
| VvAKR02 | C1630H2584N452O480S15 | 37.84 | 6.01 | 15.32 | 40.84 | Nucleus,cytosol,cytoskeleton and chloroplast |
| VvAKR03 | C1437H2314N388O409S16 | 41.32 | 4.51 | 18.06 | 36.11 | Nucleus,membrane,cytosol,chloroplast and mitochondrion,peroxisome |
| VvAKR04 | C1533H2418N402O437S17 | 42.57 | 4.62 | 13.86 | 38.94 | Nucleus,cytosol,cytoskeleton and chloroplast |
| VvAKR05 | C1584H2534N424O466S15 | 43.04 | 3.80 | 15.51 | 37.66 | Extracellular,cytosol,chloroplast,mitochondrion and endoplasmic reticulum |
| VvAKR06 | C2207H3481N571O627S19 | 34.42 | 3.95 | 17.44 | 44.19 | Nucleus,membrane,cytosol,cytoskeleton,chloroplast and mitochondrion |
| VvAKR07 | C1560H2445N401O445S12 | 41.39 | 5.30 | 13.58 | 39.74 | Nucleus,extracellular,cytosol and cytoskeleton |
| VvAKR08 | C1829H2900N490O533S14 | 42.15 | 4.41 | 13.50 | 39.94 | Nucleus,membrane,cytosol and cytoskeleton |
| VvAKR09 | C1601H2518N442O469S16 | 40.37 | 8.39 | 14.60 | 36.65 | Nucleus,membrane,cytosol,cytoskeleton and chloroplast |
| VvAKR10 | C1582H2451N419O463S10 | 42.54 | 6.67 | 13.97 | 36.83 | Nucleus,cytosol and chloroplast |
| VvAKR11 | C1378H2147N363O395S10 | 43.27 | 5.09 | 14.18 | 37.45 | Nucleus,cytosol,cytoskeleton,endoplasmic reticulum and vacuole |
| VvAKR12 | C1518H2375N411O435S10 | 43.00 | 6.67 | 12.00 | 38.33 | Nucleus,membrane,cytosol,cytoskeleton and vacuole |
| VvAKR013 | C1872H2916N492O531S11 | 48.91 | 6.25 | 11.41 | 33.42 | Nucleus,cytosol and chloroplast |
| [1] |
Mittler R. Oxidative stress, antioxidants and stress tolerance[J]. Trends Plant Sci, 2002, 7(9):405-410.
pmid: 12234732 |
| [2] |
Hegedüs A, Erdei S, Janda T, et al. Transgenic tobacco plants overproducing alfalfa aldose/aldehyde reductase show higher tolerance to low temperature and cadmium stress[J]. Plant Sci, 2004, 166(5):1329-1333.
doi: 10.1016/j.plantsci.2004.01.013 URL |
| [3] |
Simpson PJ, Tantitadapitak C, Reed AM, et al. Characterization of two novel aldo-keto reductases from Arabidopsis:expression patterns, broad substrate specificity, and an open active-site structure suggest a role in toxicant metabolism following stress[J]. J Mol Biol, 2009, 392(2):465-480.
doi: 10.1016/j.jmb.2009.07.023 pmid: 19616008 |
| [4] |
Spite M, Baba SP, Ahmed Y, et al. Substrate specificity and catalytic efficiency of aldo-keto reductases with phospholipid aldehydes[J]. Biochem J, 2007, 405(1):95-105.
doi: 10.1042/BJ20061743 URL |
| [5] |
Lee EH, Song DG, Lee JY, et al. Inhibitory effect of the compounds isolated from Rhus verniciflua on aldose reductase and advanced glycation endproducts[J]. Biol Pharm Bull, 2008, 31(8):1626-1630.
doi: 10.1248/bpb.31.1626 URL |
| [6] |
Mudalkar S, Sreeharsha RV, Reddy AR. A novel aldo-keto reductase from Jatropha curcas L. (JcAKR)plays a crucial role in the detoxification of methylglyoxal, a potent electrophile[J]. J Plant Physiol, 2016, 195:39-49.
doi: 10.1016/j.jplph.2016.03.005 URL |
| [7] | Éva C, Zelenyánszki H, Tömösközi-Farkas R, et al. Transgenic barley expressing the Arabidopsis AKR4C9 aldo-keto reductase enzyme exhibits enhanced freezing tolerance and regenerative capacity[J]. S Afr N J Bot, 2014, 93:179-184. |
| [8] | 蔡秋香, 廖端芳, 祖旭宇, 等. 醛酮还原酶超家族的研究进展[J]. 医学综述, 2008, 14(24):3693-3695. |
| Cai QX, Liao DF, Zu XY, et al. Research trends of alketose-reductase[J]. Med Recapitul, 2008, 14(24):3693-3695. | |
| [9] |
Gallego O, Ruiz FX, Ardèvol A, et al. Structural basis for the high all-trans-retinaldehyde reductase activity of the tumor marker AKR1B10[J]. PNAS, 2007, 104(52):20764-20769.
pmid: 18087047 |
| [10] |
Jez JM, Bennett MJ, Schlegel BP, et al. Comparative anatomy of the aldo-keto reductase superfamily[J]. Biochem J, 1997, 326(Pt 3):625-636.
doi: 10.1042/bj3260625 URL |
| [11] |
Petschacher B, Leitgeb S, Kavanagh KL, et al. The coenzyme specificity of Candida tenuis xylose reductase(AKR2B5)explored by site-directed mutagenesis and X-ray crystallography[J]. Biochem J, 2005, 385(pt 1):75-83.
pmid: 15320875 |
| [12] |
Alban C, Baldet P, Douce R. Localization and characterization of two structurally different forms of acetyl-CoA carboxylase in young pea leaves, of which one is sensitive to aryloxyphenoxypropionate herbicides[J]. Biochem J, 1994, 300(Pt 2):557-565.
doi: 10.1042/bj3000557 URL |
| [13] |
Sengupta D, Naik D, Reddy AR. Plant aldo-keto reductases (AKRs)as multi-tasking soldiers involved in diverse plant metabolic processes and stress defense:a structure-function update[J]. J Plant Physiol, 2015, 179:40-55.
doi: 10.1016/j.jplph.2015.03.004 URL |
| [14] |
Éva C, Tóth G, Oszvald M, et al. Overproduction of an Arabidopsis aldo-keto reductase increases barley tolerance to oxidative and cadmium stress by an in vivo reactive aldehyde detoxification[J]. Plant Growth Regul, 2014, 74(1):55-63.
doi: 10.1007/s10725-014-9896-x URL |
| [15] |
Suekawa M, Fujikawa Y, et al. Gene expression and promoter analysis of a novel tomato aldo-keto reductase in response to environmental stresses[J]. J Plant Physiol, 2016, 200:35-44.
doi: 10.1016/j.jplph.2016.05.015 URL |
| [16] | 李立威. 甘蓝醛还原酶基因克隆及其功能分析[D]. 福州:福建农林大学, 2014. |
| Li LW. Cloning and functional analysis of aldehyde reductase gene in Brassica oleracea[D]. Fuzhou:Fujian Agriculture and Forestry University, 2014. | |
| [17] | 张飞雪, 虞章红, 刘坤宇, 等. 不结球白菜醛酮还原酶BcAKR4C9基因的克隆及表达分析[J]. 南京农业大学学报, 2018, 41(2):240-247. |
| Zhang FX, Yu ZH, Liu KY, et al. Cloning and expression analysis of aldo-keto reductase gene BcAKR4C9 from non-heading Chinese cabbage[J]. J Nanjing Agric Univ, 2018, 41(2):240-247. | |
| [18] | 杨洋, 李鸿彬. 棉纤维醛酮还原酶基因促进拟南芥主根伸长[J]. 石河子大学学报:自然科学版, 2017, 35(5):612-617. |
| Yang Y, Li HB. A cotton fiber aldehyde ketone reductase gene Gh AKR promotes elongation of main root in Arabidopsis thaliana[J]. J Shihezi Univ:Nat Sci, 2017, 35(5):612-617. | |
| [19] |
Cai XF, Zhang CJ, Ye J, et al. Ectopic expression of FaGalUR leads to ascorbate accumulation with enhanced oxidative stress, cold, and salt tolerance in tomato[J]. Plant Growth Regul, 2015, 76(2):187-197.
doi: 10.1007/s10725-014-9988-7 URL |
| [20] |
Kanayama Y, Mizutani R, Yaguchi S, et al. Characterization of an uncharacterized aldo-keto reductase gene from peach and its role in abiotic stress tolerance[J]. Phytochemistry, 2014, 104:30-36.
doi: 10.1016/j.phytochem.2014.04.008 URL |
| [21] |
Wilkins MR, Gasteiger E, Bairoch A, et al. Protein identification and analysis tools in the ExPASy server[J]. Methods Mol Biol, 1999, 112:531-552.
pmid: 10027275 |
| [22] |
Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2. 0[J]. Bioinform Oxf Engl, 2007, 23(21):2947-2948.
doi: 10.1093/bioinformatics/btm404 URL |
| [23] |
Kumar S, Stecher G, Tamura K. MEGA7:molecular evolutionary genetics analysis version 7. 0 for bigger datasets[J]. Mol Biol Evol, 2016, 33(7):1870-1874.
doi: 10.1093/molbev/msw054 URL |
| [24] |
Zhang ZB, Zhang JW, Chen YJ, et al. Genome-wide analysis and identification of HAK potassium transporter gene family in maize(Zea mays L.)[J]. Mol Biol Rep, 2012, 39(8):8465-8473.
doi: 10.1007/s11033-012-1700-2 URL |
| [25] |
Bailey TL, Boden M, Buske FA, et al. MEME SUITE:tools for motif discovery and searching[J]. Nucleic Acids Res, 2009, 37(web server issue):W202-W208.
doi: 10.1093/nar/gkp335 URL |
| [26] |
Higo K, Ugawa Y, Iwamoto M, et al. Plant Cis-acting regulatory DNA elements(PLACE)database:1999[J]. Nucleic Acids Res, 1999, 27(1):297-300.
pmid: 9847208 |
| [27] |
Yu J, Sun H, Zhang JJ, et al. Analysis of aldo-keto reductase gene family and their responses to salt, drought, and abscisic acid stresses in Medicago truncatula[J]. Int J Mol Sci, 2020, 21(3):754.
doi: 10.3390/ijms21030754 URL |
| [28] |
Penning TM. The aldo-keto reductases(AKRs):Overview[J]. Chem Biol Interactions, 2015, 234:236-246.
doi: 10.1016/j.cbi.2014.09.024 URL |
| [29] |
Gavidia I, Pérez-Bermúdez P, Seitz HU. Cloning and expression of two novel aldo-keto reductases from Digitalis purpurea leaves[J]. Eur J Biochem, 2002, 269(12):2842-2850.
pmid: 12071946 |
| [30] |
Hideg, Nagy T, Oberschall A, et al. Detoxification function of aldose/aldehyde reductase during drought and ultraviolet-B(280-320 nm)stresses[J]. Plant Cell Environ, 2003, 26(4):513-522.
doi: 10.1046/j.1365-3040.2003.00982.x URL |
| [31] | Oberschall A, Deák M, Török K, et al. A novel aldose/aldehyde reductase protects transgenic plants against lipid peroxidation under chemical and drought stresses[J]. The Plant Journal, 2000, 24(4):437-446. |
| [32] |
Colrat S, Latché A, Guis M, et al. Purification and characterization of a NADPH-dependent aldehyde reductase from mung bean that detoxifies eutypine, a toxin from Eutypa lata1[J]. Plant Physiol, 1999, 119(2):621-626.
pmid: 9952458 |
| [33] |
Guillén P, Guis M, Martínez-Reina G, et al. A novel NADPH-dependent aldehyde reductase gene from Vigna radiata confers resistance to the grapevine fungal toxin eutypine[J]. The Plant Journal, 1998, 16(3):335-343.
doi: 10.1046/j.1365-313x.1998.00303.x URL |
| [1] | SONG Zhi-zhong, XU Wei-hua, XIAO Hui-lin, TANG Mei-ling, CHEN Jing-hui, GUAN Xue-qiang, LIU Wan-hao. Cloning, Expression and Function of Iron Regulated Transporter VvIRT1 in Wine Grape(Vitis vinifera L.) [J]. Biotechnology Bulletin, 2023, 39(8): 234-240. |
| [2] | WANG Shuai, FENG Yu-mei, BAI Miao, DU Wei-jun, YUE Ai-qin. Functional Analysis of Soybean Gene GmHMGR Responding to Exogenous Hormones and Abiotic Stresses [J]. Biotechnology Bulletin, 2023, 39(7): 131-142. |
| [3] | WEI Xi-ya, QIN Zhong-wei, LIANG La-mei, LIN Xin-qi, LI Ying-zhi. Mechanism of Melatonin Seed Priming in Improving Salt Tolerance of Capsicum annuum [J]. Biotechnology Bulletin, 2023, 39(7): 160-172. |
| [4] | ZHANG Lu-yang, HAN Wen-long, XU Xiao-wen, YAO Jian, LI Fang-fang, TIAN Xiao-yuan, ZHANG Zhi-qiang. Identification and Expression Analysis of the Tobacco TCP Gene Family [J]. Biotechnology Bulletin, 2023, 39(6): 248-258. |
| [5] | LI Jing-rui, WANG Yu-bo, XIE Zi-wei, LI Chang, WU Xiao-lei, GONG Bin-bin, GAO Hong-bo. Identification and Expression Analysis of PIN Gene Family in Melon Under High Temperature Stress [J]. Biotechnology Bulletin, 2023, 39(5): 192-204. |
| [6] | GUO San-bao, SONG Mei-ling, LI Ling-xin, YAO Zi-zhao, GUI Ming-ming, HUANG Sheng-he. Cloning and Analysis of Chalcone Synthase Gene and Its Promoter from Euphorbia maculata [J]. Biotechnology Bulletin, 2023, 39(4): 148-156. |
| [7] | WANG Yi-qing, WANG Tao, WEI Chao-ling, DAI Hao-min, CAO Shi-xian, SUN Wei-jiang, ZENG Wen. Identification and Interaction Analysis of SMAS Gene Family in Tea Plant(Camellia sinensis) [J]. Biotechnology Bulletin, 2023, 39(4): 246-258. |
| [8] | YANG Lan, ZHANG Chen-xi, FAN Xue-wei, WANG Yang-guang, WANG Chun-xiu, LI Wen-ting. Gene Cloning, Expression Pattern, and Promoter Activity Analysis of Chicken BMP15 [J]. Biotechnology Bulletin, 2023, 39(4): 304-312. |
| [9] | WANG Hai-long, LI Yu-qian, WANG Bo, XING Guo-fang, ZHANG Jie-wei. Isolation and Expression Analysis of SiMAPK3 in Setaria italica L. [J]. Biotechnology Bulletin, 2023, 39(3): 123-132. |
| [10] | CHEN Qiang, ZHOU Ming-kang, SONG Jia-min, ZHANG Chong, WU Long-kun. Identification and Analysis of LBD Gene Family and Expression Analysis of Fruit Development in Cucumis melo [J]. Biotechnology Bulletin, 2023, 39(3): 176-183. |
| [11] | PING Huai-lei, GUO Xue, YU Xiao, SONG Jing, DU Chun, WANG Juan, ZHANG Huai-bi. Cloning and Expression of PdANS in Paeonia delavayi and Correlation with Anthocyanin Content [J]. Biotechnology Bulletin, 2023, 39(3): 206-217. |
| [12] | DU Qing-jie, ZHOU Lu-yao, YANG Si-zhen, ZHANG Jia-xin, CHEN Chun-lin, LI Juan-qi, LI Meng, ZHAO Shi-wen, XIAO Huai-juan, WANG Ji-qing. Overexpression of CaCP1 Enhances Salt Stress Sensibility in Transgenic Tobacco [J]. Biotechnology Bulletin, 2023, 39(2): 172-182. |
| [13] | WANG Ming-tao, LIU Jian-wei, ZHAO Chun-zhao. Molecular Mechanisms of Cell Wall Integrity in Plants Under Salt Stress [J]. Biotechnology Bulletin, 2023, 39(11): 18-27. |
| [14] | XING Yuan, SONG Jian, LI Jun-yi, ZHENG Ting-ting, LIU Si-chen, QIAO Zhi-jun. Identification of AP Gene Family and Its Response Analysis to Abiotic Stress in Setaria italica [J]. Biotechnology Bulletin, 2023, 39(11): 238-251. |
| [15] | CHEN Chu-yi, YANG Xiao-mei, CHEN Sheng-yan, CHEN Bin, YUE Li-ran. Expression Analysis of the ZF-HD Gene Family in Chrysanthemum nankingense Under Drought and ABA Treatment [J]. Biotechnology Bulletin, 2023, 39(11): 270-282. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||