








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (1): 70-76.doi: 10.13560/j.cnki.biotech.bull.1985.2021-0326
Previous Articles Next Articles
Received:2021-03-16
Online:2022-01-26
Published:2022-02-22
Contact:
HAN Lie-bao
E-mail:raphael_ryan@163.com;hanliebao@163.com
WANG Rui, HAN Lie-bao. Generation of bdfls2-knockout Mutant in Brachypodium distachyon Mediated by CRISPR/Cas9[J]. Biotechnology Bulletin, 2022, 38(1): 70-76.
| 组分 Component | 体积 Volume/μL | 反应条件 Reaction conditions |
|---|---|---|
| 退火后的靶点引物 | 2 | 5 h at 37℃ 5 min at 50℃ 10 min at 80℃ |
| pHUE411 载体(约100 ng/μL) | 2 | |
| 10× T4 DNA Ligase Buffer(NEB) | 1.5 | |
| 10× BSA | 1.5 | |
| Bsa I(NEB) | 1 | |
| T4 DNA Ligase(NEB) | 1 | |
| ddH2O | 6 |
Table 1 Enzyme digestion-connection system
| 组分 Component | 体积 Volume/μL | 反应条件 Reaction conditions |
|---|---|---|
| 退火后的靶点引物 | 2 | 5 h at 37℃ 5 min at 50℃ 10 min at 80℃ |
| pHUE411 载体(约100 ng/μL) | 2 | |
| 10× T4 DNA Ligase Buffer(NEB) | 1.5 | |
| 10× BSA | 1.5 | |
| Bsa I(NEB) | 1 | |
| T4 DNA Ligase(NEB) | 1 | |
| ddH2O | 6 |
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| BdPR2 -F | AGCCATCCAGCTCAACTAC |
| BdPR2 -R | CCTTGCCAACATGGTCAATC |
| BdChit8 -F | CTGCTTCAAGGAGGAGATAAAC |
| BdChit8 -R | TCATCCAGAACCACATGGC |
| BdAct -F | GCTGGGCGTGACCTAACTGAC |
| BdAct -R | ATGAAAGATGGCTGGAAAAGGACT |
Table 2 Sequence of primer for qRT-PCR
| 引物名称Primer name | 引物序列Primer sequence(5'-3') |
|---|---|
| BdPR2 -F | AGCCATCCAGCTCAACTAC |
| BdPR2 -R | CCTTGCCAACATGGTCAATC |
| BdChit8 -F | CTGCTTCAAGGAGGAGATAAAC |
| BdChit8 -R | TCATCCAGAACCACATGGC |
| BdAct -F | GCTGGGCGTGACCTAACTGAC |
| BdAct -R | ATGAAAGATGGCTGGAAAAGGACT |
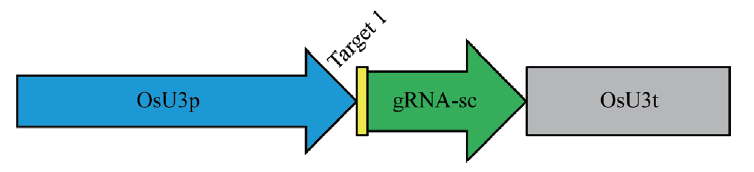
Fig. 1 Illustration of CRISPR/Cas9-bdfls2 gene editing vector in B. distachyon OsU3:sgRNA-Cas9-pHUE411 vector. Light blue indicates the Oryzae U3 promoter. Yellow and green portions indicate the sgRNA sequence. The yellow portion indicates the target sequence of the gene. The grey portion indicates the Oryzae U3 terminator
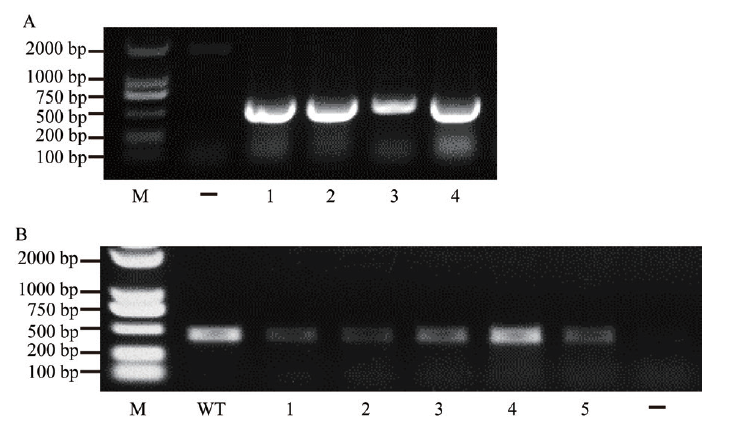
Fig. 2 Identification of gene editing vector and edited target gene A:Identification of CRISPR/Cas9-bdfls2 gene editing vector. 1-4 are the assayed Agrobacterium tumefaciens strains for transformed CRISPR/Cas9-bdfls2. B:Amplification and identification of target gene. 1-5 are genome-DNA-amplified fragments containing target gene editing sites extracted from transgenic plants. M is the DNA marker. Bd21 is the wild type(WT). - indicates the negative control
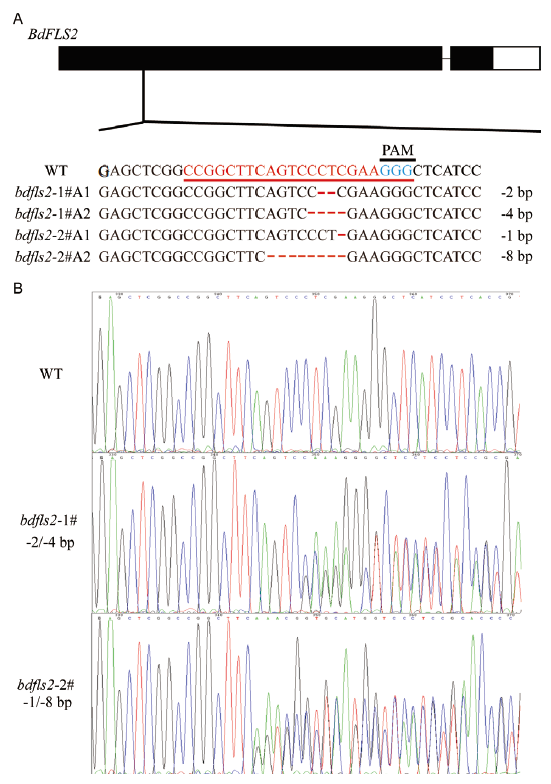
Fig. 3 Genotyping of bdfls2 mutants in B. distachyon A:Genotyping of bdfls2 mutants. Red underline refers to sgRNA:Cas9-edited bdfls2 mutation. Blue underline refer to schematic illustration of the sgRNA:Cas9 targets and the corresponding protospacer-adjacent motif(PAM)(blue)of the targeted genes. The 1# and 2# stands for different mutant plants,A1 and A2 for Allele 1 and Allele 2,and the red lines in sequences stands for base deletion. B:Sequencing of bdfls2 mutants. WT is the Bd21 wild type as control
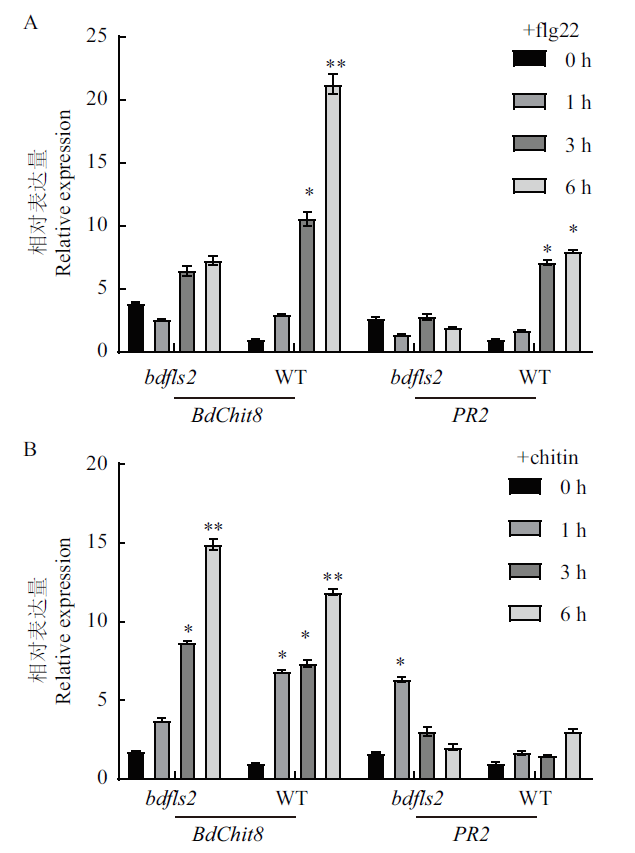
Fig. 5 Detection of defense gene expression induced upon different PAMPs treatment by real-time quantitative RT-PCR The relative BdChit8 and PR2 expression in the leaves of B. distachyon seedlings treated with 5 μmol/L flg22(A)or 100 μg/mL chitin(B)were determined by quantitative real-time PCR. Data are presented as the mean ± standard error of three biological replicates.* or ** above the bars indicate P < 0.05 or P < 0.01
| [1] |
Ausubel FM. Are innate immune signaling pathways in plants and animals conserved?[J]. Nat Immunol, 2005, 6(10):973-979.
doi: 10.1038/ni1253 URL |
| [2] |
Jones JDG, Dangl JL. The plant immune system[J]. Nature, 2006, 444(7117):323-329.
doi: 10.1038/nature05286 URL |
| [3] |
Sanabria N, Goring D, Nürnberger T, et al. Self/nonself perception and recognition mechanisms in plants:a comparison of self-incompatibility and innate immunity[J]. New Phytol, 2008, 178(3):503-514.
doi: 10.1111/j.1469-8137.2008.02403.x pmid: 18346103 |
| [4] |
Zipfel C. Plant pattern-recognition receptors[J]. Trends Immunol, 2014, 35(7):345-351.
doi: 10.1016/j.it.2014.05.004 pmid: 24946686 |
| [5] |
Bigeard J, Colcombet J, Hirt H. Signaling mechanisms in pattern-triggered immunity(PTI)[J]. Mol Plant, 2015, 8(4):521-539.
doi: 10.1016/j.molp.2014.12.022 pmid: 25744358 |
| [6] |
Zipfel C. Pattern-recognition receptors in plant innate immunity[J]. Curr Opin Immunol, 2008, 20(1):10-16.
doi: 10.1016/j.coi.2007.11.003 pmid: 18206360 |
| [7] |
Gómez-Gómez L, Felix G, Boller T. A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana[J]. Plant J, 1999, 18(3):277-284.
pmid: 10377993 |
| [8] |
Robatzek S, Bittel P, Chinchilla D, et al. Molecular identification and characterization of the tomato flagellin receptor LeFLS2, an orthologue of Arabidopsis FLS2 exhibiting characteristically different perception specificities[J]. Plant Mol Biol, 2007, 64(5):539-547.
pmid: 17530419 |
| [9] |
Huang PY, Yeh YH, Liu AC, et al. The Arabidopsis LecRK-VI. 2 associates with the pattern-recognition receptor FLS2 and primes Nicotiana benthamiana pattern-triggered immunity[J]. Plant J, 2014, 79(2):243-255.
doi: 10.1111/tpj.12557 URL |
| [10] |
Gómez-Gómez L, Boller T. FLS2:an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis[J]. Mol Cell, 2000, 5(6):1003-1011.
pmid: 10911994 |
| [11] |
Takai R, Isogai A, Takayama S, et al. Analysis of flagellin perception mediated by flg22 receptor OsFLS2 in rice[J]. Mol Plant Microbe Interact, 2008, 21(12):1635-1642.
doi: 10.1094/MPMI-21-12-1635 URL |
| [12] |
Hann DR, Rathjen JP. Early events in the pathogenicity of Pseudomonas syringae on Nicotiana benthamiana[J]. Plant J, 2007, 49(4):607-618.
doi: 10.1111/j.1365-313X.2006.02981.x URL |
| [13] |
Boller T, Felix G. A renaissance of elicitors:perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors[J]. Annu Rev Plant Biol, 2009, 60:379-406.
doi: 10.1146/arplant.2009.60.issue-1 URL |
| [14] |
Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and Archaea[J]. Nature, 2012, 482(7385):331-338.
doi: 10.1038/nature10886 URL |
| [15] |
Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering[J]. Cell, 2014, 157(6):1262-1278.
doi: 10.1016/j.cell.2014.05.010 URL |
| [16] |
Yin K, Gao C, Qiu JL. Progress and prospects in plant genome editing[J]. Nat Plants, 2017, 3:17107.
doi: 10.1038/nplants.2017.107 URL |
| [17] |
Ji X, Wang D, Gao C. CRISPR editing-mediated antiviral immunity:a versatile source of resistance to combat plant virus infections[J]. Sci China Life Sci, 2019, 62(9):1246-1249.
doi: 10.1007/s11427-019-9722-2 URL |
| [18] |
Kellogg EA. Evolutionary history of the grasses[J]. Plant Physiol, 2001, 125(3):1198-1205.
pmid: 11244101 |
| [19] |
Davis JI, Soreng RJ. Phylogenetic structure in the grass family(Poaceae)as inferred from chloroplast DNA restriction site variation[J]. Am J Bot, 1993, 80(12):1444.
doi: 10.1002/ajb2.1993.80.issue-12 URL |
| [20] |
Liu H, Ding Y, Zhou Y, et al. CRISPR-P 2. 0:an improved CRISPR-Cas9 tool for genome editing in plants[J]. Mol Plant, 2017, 10(3):530-532.
doi: 10.1016/j.molp.2017.01.003 URL |
| [21] |
Chinchilla D, Bauer Z, Regenass M, et al. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception[J]. Plant Cell, 2006, 18(2):465-476.
pmid: 16377758 |
| [22] |
Xing HL, Dong L, Wang ZP, et al. A CRISPR/Cas9 toolkit for multiplex genome editing in plants[J]. BMC Plant Biol, 2014, 14:327.
doi: 10.1186/s12870-014-0327-y URL |
| [23] | 吴雪莉, 刘金星, Nielsen K, 等. 二穗短柄草幼胚再生体系及农杆菌介导转化的初步研究[J]. 草业学报, 2010, 19(5):9-16. |
| Wu XL, Liu JX, Nielsen K, et al. Agrobacterium-mediated transformation of Brachypodium distachyon through embryogenic calli derived from immature embryos[J]. Acta Prataculturae Sin, 2010, 19(5):9-16. | |
| [24] |
Guo W, Zuo ZL, Cheng X, et al. The chloride channel family gene CLCd negatively regulates pathogen-associated molecular pattern(PAMP)-triggered immunity in Arabidopsis[J]. J Exp Bot, 2014, 65(4):1205-1215.
doi: 10.1093/jxb/ert484 URL |
| [25] |
Pérez FJ, Rubio S. An improved chemiluminescence method for hydrogen peroxide determination in plant tissues[J]. Plant Growth Regul, 2006, 48(1):89-95.
doi: 10.1007/s10725-005-5089-y URL |
| [26] |
Liu W, Xie X, Ma X, et al. DSDecode:a web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations[J]. Mol Plant, 2015, 8(9):1431-1433.
doi: 10.1016/j.molp.2015.05.009 URL |
| [27] |
Wang S, Sun Z, Wang H, et al. Rice OsFLS2-mediated perception of bacterial flagellins is evaded by Xanthomonas oryzae pvs. oryzae and oryzicola[J]. Mol Plant, 2015, 8(7):1024-1037.
doi: 10.1016/j.molp.2015.01.012 URL |
| [28] |
Fürst U, Zeng Y, Albert M, et al. Perception of Agrobacterium tumefaciens flagellin by FLS2XL confers resistance to crown gall disease[J]. Nat Plants, 2020, 6(1):22-27.
doi: 10.1038/s41477-019-0578-6 URL |
| [1] | CHEN Xiao-ling, LIAO Dong-qing, HUANG Shang-fei, CHEN Ying, LU Zhi-long, CHEN Dong. Advances in CRISPR/Cas9 System Modifying Saccharomycescerevisiae [J]. Biotechnology Bulletin, 2023, 39(8): 148-158. |
| [2] | YANG Yu-mei, ZHANG Kun-xiao. Establishing a Stable Cell Line with Site-specific Integration of ERK Kinase Phase-separated Fluorescent Probe Using CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(8): 159-164. |
| [3] | SHI Wei-tao, YAO Chun-peng, WEI Wen-Kang, WANG Lei, FANG Yuan-jie, TONG Yu-jie, MA Xiao-jiao, JIANG Wen, ZHANG Xiao-ai, SHAO Wei. Establishment of MDH2 Knockout Cell Line Using CRISPR/Cas9 Technology and Study of Anti-deoxynivalenol Effect [J]. Biotechnology Bulletin, 2023, 39(7): 307-315. |
| [4] | LIU Xiao-yan, ZHU Zhen-liang, SHI Guang-yu, HUA Zi-yu, YANG Chen, ZHANG Yong, LIU Jun. Strategies to Optimize the Expression of Mammary Gland Bioreactor [J]. Biotechnology Bulletin, 2023, 39(5): 77-91. |
| [5] | CHENG Jing-wen, CAO Lei, ZHANG Yan-min, YE Qian, CHEN Min, TAN Wen-song, ZHAO Liang. Establishment and Application of Multigene Engineering Transformation Strategy for CHO Cells [J]. Biotechnology Bulletin, 2023, 39(2): 283-291. |
| [6] | HUANG Wen-li, LI Xiang-xiang, ZHOU Wen-ting, LUO Sha, YAO Wei-jia, MA Jie, ZHANG Fen, SHEN Yu-sen, GU Hong-hui, WANG Jian-sheng, SUN Bo. Targeted Editing of BoZDS in Broccoli by CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2023, 39(2): 80-87. |
| [7] | WANG Bing, ZHAO Hui-na, YU Jing, CHEN Jie, LUO Mei, LEI Bo. Regulation of Leaf Bud by REVOLUTA in Tobacco Based on CRISPR/Cas9 System [J]. Biotechnology Bulletin, 2023, 39(10): 197-208. |
| [8] | LI Shuang-xi, HUA Jin-lian. Research Progress in Anti-porcine Reproductive and Respiratory Syndrome Genetically Modified Pigs [J]. Biotechnology Bulletin, 2023, 39(10): 50-57. |
| [9] | LIN Rong, ZHENG Yue-ping, XU Xue-zhen, LI Dan-dan, ZHENG Zhi-fu. Functional Analysis of ACOL8 Gene in the Ethylene Synthesis and Response in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2023, 39(1): 157-165. |
| [10] | LIU Jing-jing, LIU Xiao-rui, LI Lin, WANG Ying, YANG Hai-yuan, DAI Yi-fan. Establishment of Porcine Fetal Fibroblasts with OXTR-knockout Using CRISPR/Cas9 [J]. Biotechnology Bulletin, 2022, 38(6): 272-278. |
| [11] | Olalekan Amoo, HU Li-min, ZHAI Yun-gu, FAN Chu-chuan, ZHOU Yong-ming. Regulation of Shoot Branching by BRANCHED1 in Brassica napus Based on Gene Editing Technology [J]. Biotechnology Bulletin, 2022, 38(4): 97-105. |
| [12] | DING Ya-qun, DING Ning, XIE Shen-min, HUANG Meng-na, ZHANG Yu, ZHANG Qin, JIANG Li. Construction of Vps28 Knock-out Mice and Model Study of the Impact on Lactation and Immune Traits [J]. Biotechnology Bulletin, 2022, 38(3): 164-172. |
| [13] | YAN Jiong, FENG Chen-yi, GAO Xue-kun, XU Xiang, YANG Jia-min, CHEN Zhao-yang. Construction of Homozygous Plin1-knockout Mouse Model and Phenotype Analysis Based on CRISPR/Cas9 Technology [J]. Biotechnology Bulletin, 2022, 38(3): 173-180. |
| [14] | ZHONG Jing, SUN Ling-ling, ZHANG Shu, MENG Yuan, ZHI Yi-fei, TU Li-qing, XU Tian-peng, PU Li-ping, LU Yang-qing. Effect of Knocking Out the Mda5 Gene by CRISPR/Cas9 Technology on the Replication of Newcastle Disease and Infectious Bursal Virus [J]. Biotechnology Bulletin, 2022, 38(11): 90-96. |
| [15] | ZONG Mei, HAN Shuo, GUO Ning, DUAN Meng-meng, LIU Fan, WANG Gui-xiang. Production of Marker-free Mutants of Brassica campestris Mediated by CRISPR/Cas9 Through Vacuum Infiltration [J]. Biotechnology Bulletin, 2022, 38(10): 159-163. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||
