








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (10): 34-44.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1558
Previous Articles Next Articles
SHAN Qi1( ), JIA Hui-shu1, YAO Wen-bo1, LIU Wei-can1(
), JIA Hui-shu1, YAO Wen-bo1, LIU Wei-can1( ), LI Hai-yan2(
), LI Hai-yan2( )
)
Received:2021-12-16
Online:2022-10-26
Published:2022-11-11
Contact:
LIU Wei-can,LI Hai-yan
E-mail:SQ1806006978@163.com;liuweican602@163.com;hyli99@163.com
SHAN Qi, JIA Hui-shu, YAO Wen-bo, LIU Wei-can, LI Hai-yan. Research Progress in Biological Functions of miR396-GRF Module in Plants and Its Potential Application Values[J]. Biotechnology Bulletin, 2022, 38(10): 34-44.

Fig.1 Alignment analysis of mature region sequences of miR396 gene family in different plant species A and B:The most mat-miR396 sequences in plants are mainly the same as the 2 mature sequence forms of ath-miR396a-5p and ath-miR396b-5p in Arabidopsis,ath-miR396a-5p and ath-miR396b-3p have the same sequence length,but they have a different base at the end. C:The alignment of other mat-miR396 sequences except above 2 sequences in plants revealed that there is one base difference or multiple base differences between mat-miR396 sequences

Fig.2 Alignment analysis of the conserved domains of GRF protein family amino acid sequences in different plant species According to the amino acid sequence comparison analysis of GRF protein family in different plant species,it can be found that the N-terminal of GRF protein contained two conservative protein domain,QLQ and WRC. A:QLQ domain of GRF protein. In the QLQ domain,there are conserved Gln-Leu-GLn(Q-L-Q)amino acids and hydrophobic residues such as F/Phe,Y/Tyr,L/Leu,E/Glu and P/Pro,or other amino acids with similar physical and chemical properties. B:WRC domain of GRF protein. In the WRC domain,there are conserved Trp-Arg-Cys(W-R-C)amino acid residues and C3H motifs consisting of three C/Cys and an H/His. It is important to note that there is a conserved “RSRK-VE” region following the C3H motif region,its coded nucleotide sequence is the targets site of miR396 on the GRF gene

Fig.3 Action sites and splicing sites of miR396 on GRF gene sequences in different plant species A:The action site of miR396 on GRF gene. In the WRC domain of GRF protein,there is a conserved “RSRK-VE ”region,their corresponding nucleotide sequences in the genome are mostly highly conserved “CGTTCAAGAACNTGTGAA” sequences. B:Splice site of miR396 on the mRNA sequence of GRF gene. miR396 cut the mRNA sequence of GRF gene between “CGUUCAAGAA” and “AGCNUGUGGAA” site,thus negatively regulating the GRF gene expression
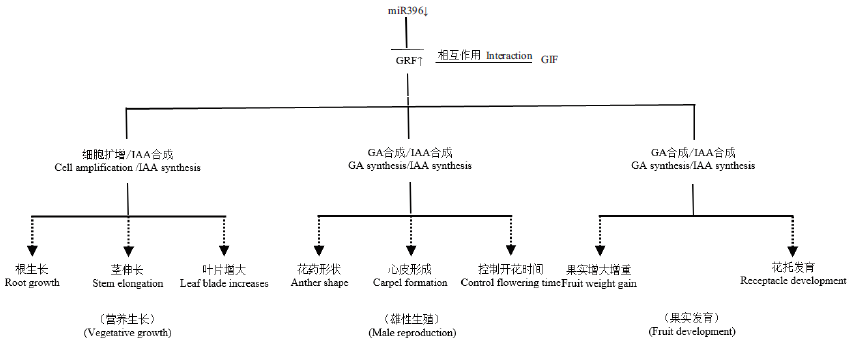
Fig. 4 Process of miR396-GRF module regulating plant growth and development The miR396-GRF regulatory pathway affects vegetative growth,male reproduction and fruit development by affecting cell amplification and the synthesis of GA and IAA hormones. And GIF protein and GRF protein will form a functional complex to enhance GRF function
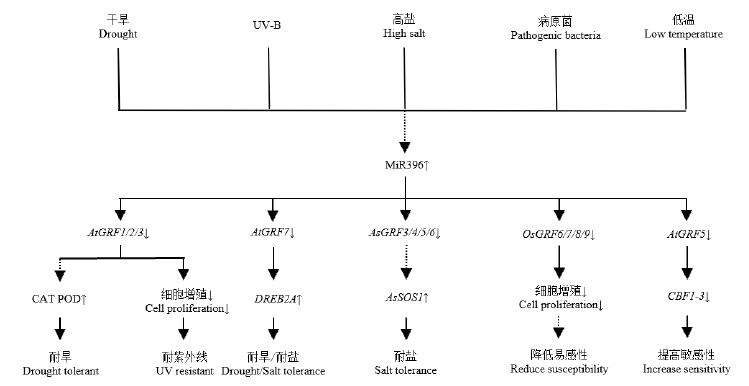
Fig. 5 Feasible regulation approach of miR396-GRF mod-ule in the process of stress response In plants, miR396-GRF module participates in a variety of stress response processes by regulating the expression of downstream stress response related functional genes and cell proliferation
| [1] |
Liu WC, Zhou YG, Li XW, et al. Tissue-specific regulation of gma-miR396 family on coordinating development and low water availability responses[J]. Front Plant Sci, 2017, 8:1112.
doi: 10.3389/fpls.2017.01112 pmid: 28694817 |
| [2] |
Omidbakhshfard MA, Proost S, Fujikura U, et al. Growth-regulating factors(GRFs):a small transcription factor family with important functions in plant biology[J]. Mol Plant, 2015, 8(7):998-1010.
doi: 10.1016/j.molp.2015.01.013 pmid: 25620770 |
| [3] |
Chen XL, Jiang LR, Zheng JS, et al. A missense mutation in Large Grain Size 1 increases grain size and enhances cold tolerance in rice[J]. J Exp Bot, 2019, 70(15):3851-3866.
doi: 10.1093/jxb/erz192 pmid: 31020332 |
| [4] | 强晓敏, 高南, 冯晓宇, 等. 超量表达GRF9基因促进番茄生长并增强其对磷的吸收利用能力[J]. 土壤, 2013, 45(3):483-488. |
| Qiang XM, Gao N, Feng XY, et al. GRF9 over-expressing improves tomato growth and phosphorus use efficiency[J]. Soils, 2013, 45(3):483-488. | |
| [5] |
韩美玲, 谭茹姣, 晁代印. “绿色革命”新进展:赤霉素与氮营养双重调控的表观修饰助力水稻高产高效育种[J]. 植物学报, 2020, 55(1):5-8.
doi: 10.11983/CBB20002 |
| Han ML, Tan RJ, Chao DY. A new progress of green revolution:epigenetic modification dual-regulated by gibberellin and nitrogen supply contributes to breeding of high yield and nitrogen use efficiency rice[J]. Chin Bull Bot, 2020, 55(1):5-8. | |
| [6] |
Kong JX, Martin-Ortigosa S, Finer J, et al. Overexpression of the transcription factor GROWTH-REGULATING FACTOR5 improves transformation of dicot and monocot species[J]. Front Plant Sci, 2020, 11:572319.
doi: 10.3389/fpls.2020.572319 URL |
| [7] |
Wu ZJ, Wang WL, Zhuang J. Developmental processes and responses to hormonal stimuli in tea plant(Camellia sinensis)leaves are controlled by GRF and GIF gene families[J]. Funct Integr Genomics, 2017, 17(5):503-512.
doi: 10.1007/s10142-017-0553-0 URL |
| [8] | Rodriguez RE, Ercoli MF, Debernardi JM, et al. Growth-regulating factors, A transcription factor family regulating more than just plant growth[M]// Gonzalez DH. Plant Transcription Factors.Amsterdam:Elsevier, 2016:269-280. |
| [9] |
Kim JH. Biological roles and an evolutionary sketch of the GRF-GIF transcriptional complex in plants[J]. BMB Rep, 2019, 52(4):227-238.
pmid: 30885290 |
| [10] |
Beltramino M, Ercoli MF, Debernardi JM, et al. Robust increase of leaf size by Arabidopsis thaliana GRF3-like transcription factors under different growth conditions[J]. Sci Rep, 2018, 8(1):13447.
doi: 10.1038/s41598-018-29859-9 pmid: 30194309 |
| [11] | 周蕾. 烟草叶片发育相关ARF和GRF家族基因的表达分析与功能验证[D]. 北京: 中国农业科学院, 2020. |
| Zhou L. Expression analysis and functional verification of ARF and GRF family genes related to tobacco leaf development[D]. Beijing: Chinese Academy of Agricultural Sciences, 2020. | |
| [12] |
Wang FD, Qiu NW, Ding Q, et al. Genome-wide identification and analysis of the growth-regulating factor family in Chinese cabbage(Brassica rapa L. ssp. pekinensis)[J]. BMC Genomics, 2014, 15(1):807.
doi: 10.1186/1471-2164-15-807 URL |
| [13] |
Lu YZ, Meng YL, Zeng J, et al. Coordination between GROWTH-REGULATING FACTOR1 and GRF-INTERACTING FACTOR1 plays a key role in regulating leaf growth in rice[J]. BMC Plant Biol, 2020, 20(1):200.
doi: 10.1186/s12870-020-02417-0 pmid: 32384927 |
| [14] |
Gao F, Wang K, Liu Y, et al. Blocking miR396 increases rice yield by shaping inflorescence architecture[J]. Nat Plants, 2015, 2:15196.
doi: 10.1038/nplants.2015.196 pmid: 27250748 |
| [15] |
Liu HH, Guo SY, Xu YY, et al. OsmiR396d-regulated OsGRFs function in floral organogenesis in rice through binding to their targets OsJMJ706 and OsCR4[J]. Plant Physiol, 2014, 165(1):160-174.
doi: 10.1104/pp.114.235564 pmid: 24596329 |
| [16] | Baucher M, Moussawi J, Vandeputte OM, et al. A role for the miR396/GRF network in specification of organ type during flower development, as supported by ectopic expression of Populus trichocarpa miR396c in transgenic tobacco[J]. Plant Biol(Stuttg), 2013, 15(5):892-898. |
| [17] |
Hou N, Cao YL, Li FY, et al. Epigenetic regulation of miR396 expression by SWR1-C and the effect of miR396 on leaf growth and developmental phase transition in Arabidopsis[J]. J Exp Bot, 2019, 70(19):5217-5229.
doi: 10.1093/jxb/erz285 URL |
| [18] | 邢媛. OsGRF1基因突变对水稻性状的影响及机制分析[D]. 扬州: 扬州大学, 2020. |
| Xing Y. Effects of OsGRF1 gene mutation on rice characters and its mechanism analysis[D]. Yangzhou: Yangzhou University, 2020. | |
| [19] | Rodriguez RE, Mecchia MA, Debernardi JM, et al. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396[J]. Dev Camb Engl, 2010, 137(1):103-112. |
| [20] |
Wang JN, Zhou HJ, Zhao YQ, et al. PagGRF12a interacts with PagGIF1b to regulate secondary xylem development through modulating PagXND1a expression in Populus alba × P. Glandulosa[J]. J Integr Plant Biol, 2021, 63(10):1683-1694.
doi: 10.1111/jipb.13102 URL |
| [21] |
Liu YR, Yan JP, Wang KX, et al. MiR396-GRF module associates with switchgrass biomass yield and feedstock quality[J]. Plant Biotechnol J, 2021, 19(8):1523-1536.
doi: 10.1111/pbi.13567 pmid: 33567151 |
| [22] |
Zhang B, Tong YN, Luo KS, et al. Identification of GROWTH-REGULATING FACTOR transcription factors in lettuce(Lactuca sativa)genome and functional analysis of LsaGRF5 in leaf size regulation[J]. BMC Plant Biol, 2021, 21(1):485.
doi: 10.1186/s12870-021-03261-6 pmid: 34688264 |
| [23] |
Bazin J, Khan GA, Combier JP, et al. miR396 affects mycorrhization and root meristem activity in the legume Medicago truncatula[J]. Plant J, 2013, 74(6):920-934.
doi: 10.1111/tpj.12178 URL |
| [24] |
Zhang JS, Zhou ZY, Bai JJ, et al. Disruption of MIR396e and MIR396f improves rice yield under nitrogen-deficient conditions[J]. Natl Sci Rev, 2020, 7(1):102-112.
doi: 10.1093/nsr/nwz142 URL |
| [25] |
Lin YR, Zhu YW, Cui YC, et al. Derepression of specific miRNA-target genes in rice using CRISPR/Cas9[J]. J Exp Bot, 2021, 72(20):7067-7077.
doi: 10.1093/jxb/erab336 URL |
| [26] |
Yu Y, Sun FY, Chen N, et al. MiR396 regulatory network and its expression during grain development in wheat[J]. Protoplasma, 2021, 258(1):103-113.
doi: 10.1007/s00709-020-01556-3 URL |
| [27] | 郭泾磊. 小麦GRF基因家族的生物信息学分析与TaGRF1和TaGRF2的功能鉴定[D]. 杨凌: 西北农林科技大学, 2020. |
| Guo JL. Bioinformatics analysis of wheat GRF gene family and functional identification of TaGRF1 and TaGRF2[D]. Yangling: Northwest A & F University, 2020. | |
| [28] | 曹东艳. miR396在番茄果实生长发育中的功能研究[D]. 北京: 中国农业大学, 2018. |
| Cao DY. Functional identification of miR396 on tomato fruit growth and development[D]. Beijing: China Agricultural University, 2018. | |
| [29] | 石文慧. 油菜高效再生体系的建立及草甘膦抗性基因EPSPs和BnGRF2的遗传转化[D]. 兰州: 甘肃农业大学, 2017. |
| Shi WH. Establishment of efficient regeneration system and genetic transformation of glyphosate resistant genes EPSPs and BnGRF2 in rape[D]. Lanzhou: Gansu Agricultural University, 2017. | |
| [30] | 张肖逢. 玉米ga20ox5矮化突变体和转ZmGRF1基因玉米的鉴定及氮素利用率研究[D]. 北京: 中国农业科学院, 2021. |
| Zhang XF. Identification and nitrogen use efficiency analysis of ga20ox5 mutant and transgenic plants overexpressing ZmGRF1[D]. Beijing: Chinese Academy of Agricultural Sciences, 2021. | |
| [31] |
Fracasso A, Vallino M, Staropoli A, et al. Increased water use efficiency in miR396-downregulated tomato plants[J]. Plant Sci, 2021, 303:110729.
doi: 10.1016/j.plantsci.2020.110729 URL |
| [32] | 章丽丽, 李光杰, 陆玉芳, 等. 拟南芥14-3--3蛋白GRF9调控番茄根系响应水分胁迫的生理机制[J]. 土壤, 2020, 52(1):74-80. |
| Zhang LL, Li GJ, Lu YF, et al. Involvement of Arabidopsis GRF9 in tomato root growth and response under polyethylene glycol induced water stress[J]. Soils, 2020, 52(1):74-80. | |
| [33] |
Yuan SR, Zhao JM, Li ZG, et al. microRNA396-mediated alteration in plant development and salinity stress response in creeping bentgrass[J]. Hortic Res, 2019, 6:48.
doi: 10.1038/s41438-019-0130-x URL |
| [34] |
Noon JB, Hewezi T, Baum TJ. Homeostasis in the soybean miRNA396-GRF network is essential for productive soybean cyst nematode infections[J]. J Exp Bot, 2019, 70(5):1653-1668.
doi: 10.1093/jxb/erz022 URL |
| [35] |
Chandran V, Wang H, Gao F, et al. miR396- OsGRFs module balances growth and rice blast disease-resistance[J]. Front Plant Sci, 2019, 9:1999.
doi: 10.3389/fpls.2018.01999 URL |
| [36] |
Pan WB, Cheng ZT, Han ZG, et al. Efficient transformation and genome editing of watermelon assisted by genes that encode developmental regulators[J]. BioRxiv, 2021. DOI:https://doi.org/10.1101/2021.11.05.467370.
doi: https://doi.org/10.1101/2021.11.05.467370 |
| [37] |
Debernardi JM, Tricoli DM, Ercoli MF, et al. A GRF-GIF chimeric protein improves the regeneration efficiency of transgenic plants[J]. Nat Biotechnol, 2020, 38(11):1274-1279.
doi: 10.1038/s41587-020-0703-0 pmid: 33046875 |
| [38] |
Qiu FT, Xing SN, Xue CX, et al. Transient expression of a TaGRF4-TaGIF1 complex stimulates wheat regeneration and improves genome editing[J]. Sci China Life Sci, 2021. DOI:https://doi.org/10.1007/s11427-021-1949-9.
doi: https://doi.org/10.1007/s11427-021-1949-9 |
| [39] |
Luo GB, Palmgren M. GRF-GIF chimeras boost plant regeneration[J]. Trends Plant Sci, 2021, 26(3):201-204.
doi: 10.1016/j.tplants.2020.12.001 pmid: 33349565 |
| [40] |
Ai G, Zhang DD, Huang R, et al. Genome-wide identification and molecular characterization of the growth-regulating factors-interacting factor gene family in tomato[J]. Genes, 2020, 11(12):1435.
doi: 10.3390/genes11121435 URL |
| [41] | Das Gupta M, Nath U. Divergence in patterns of leaf growth polarity is associated with the expression divergence of miR396[J]. Plant Cell, 2015, 27(10):2785-2799. |
| [42] |
Kuijt SJ, Greco R, Agalou A, et al. Interaction between the growth-regulating factor and knotted1-like homeobox families of transcription factors[J]. Plant Physiol, 2014, 164(4):1952-1966.
doi: 10.1104/pp.113.222836 pmid: 24532604 |
| [43] |
Blázquez MA, Nelson DC, Weijers D. Evolution of Plant Hormone Response Pathways[J]. Annu Rev Plant Biol, 2020, 71:327-353.
doi: 10.1146/annurev-arplant-050718-100309 pmid: 32017604 |
| [44] |
Park J, Lee Y, Martinoia E, et al. Plant hormone transporters:what we know and what we would like to know[J]. BMC Biol, 2017, 15(1):93.
doi: 10.1186/s12915-017-0443-x URL |
| [45] | 郭泾磊, 张程炀, 李红霞, 等. 小麦TaGRF4基因分析与功能鉴定[J]. 西北农业学报, 2020, 29(9):1317-1324. |
| Guo JL, Zhang CY, Li HX, et al. Analysis and functional identification of TaGRF4 gene in wheat[J]. Acta Agric Boreali Occidentalis Sin, 2020, 29(9):1317-1324. | |
| [46] |
Vall-Llaura N, Fernández-Cancelo P, Nativitas-Lima I, et al. ROS-scavenging-associated transcriptional and biochemical shifts during nectarine fruit development and ripening[J]. Plant Physiol Biochem, 2022, 171:38-48.
doi: 10.1016/j.plaphy.2021.12.022 URL |
| [47] |
Kim JS, Mizoi J, Kidokoro S, et al. Arabidopsis growth-regulating factor7 functions as a transcriptional repressor of abscisic acid- and osmotic stress-responsive genes, including DREB2A[J]. Plant Cell, 2012, 24(8):3393-3405.
doi: 10.1105/tpc.112.100933 URL |
| [48] |
Wu L, Zhang DF, Xue M, et al. Overexpression of the maize GRF10, an endogenous truncated growth-regulating factor protein, leads to reduction in leaf size and plant height[J]. J Integr Plant Biol, 2014, 56(11):1053-1063.
doi: 10.1111/jipb.12220 URL |
| [49] |
Chen L, Luan YS, Zhai JM. Sp-miR396a-5p acts as a stress-responsive genes regulator by conferring tolerance to abiotic stresses and susceptibility to Phytophthora nicotianae infection in transgenic tobacco[J]. Plant Cell Rep, 2015, 34(12):2013-2025.
doi: 10.1007/s00299-015-1847-0 pmid: 26242449 |
| [50] |
Yu YH, Ni ZY, Wang Y, et al. Overexpression of soybean miR169c confers increased drought stress sensitivity in transgenic Arabidopsis thaliana[J]. Plant Sci, 2019, 285:68-78.
doi: 10.1016/j.plantsci.2019.05.003 URL |
| [1] | YU Yang, LIU Tian-hai, LIU Li-xu, TANG Jie, PENG Wei-hong, CHEN Yang, TAN Hao. Study on Aerosol Microbial Community in the Production Workshop of Morel Spawn [J]. Biotechnology Bulletin, 2023, 39(5): 267-275. |
| [2] | KONG De-zhen, NIE Ying-bin, CUI Feng-juan, SANG Wei, XU Hong-jun, TIAN Xiao-ming. Research Status and Prospect of Hybrid Wheat Seed Production [J]. Biotechnology Bulletin, 2023, 39(1): 95-103. |
| [3] | LIU Tian-hai, YANG Shu-qin, LIU Fu-peng, MIAO Ren-yun, YU Yang, WU Xiang, TANG Jie, WANG Yong, PENG Wei-hong, TAN Hao. Effects of Organic Fertilizers Fermented with Wheat Straw and Chicken Manure on the Continuous Cultivation of Morchella sextelata [J]. Biotechnology Bulletin, 2022, 38(12): 263-273. |
| [4] | KONG De-zhen, NIE Ying-bin, XU Hong-jun, CUI Feng-juan, MU Pei-yuan, TIAN Xiao-ming. Effects of Blend Seeding on the Yield,Purity and Yield Advantage of F1 in Three-line Hybrid Wheat [J]. Biotechnology Bulletin, 2022, 38(10): 132-139. |
| [5] | TANG Jia-cheng, LIANG Yi-min, MA Jia-si, PENG Gui-xiang, TAN Zhi-yuan. Diversity and Growth Promotion of Endophytic Bacteria Isolated from Passiflora edulia Sims [J]. Biotechnology Bulletin, 2022, 38(1): 86-97. |
| [6] | LIAO Zhao-min, CAI Jun, LIN Jian-guo, DU Xin, WANG Chang-gao. Expression of Glucose Oxidase Gene from Aspergillus niger in Pichia pastoris and Optimization of Enzyme Production Conditions [J]. Biotechnology Bulletin, 2021, 37(6): 97-107. |
| [7] | XU Zi-han, LIU Qian, MIAO Da-peng, CHEN Yue, HU Feng-rong. Impacts of Cymbidium goeringii’s miR396 Overexpression on the Leaf Growth,Photosynthesis and Chlorophyll Fluorescence in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2021, 37(5): 28-37. |
| [8] | TAO Zhi-dong, HE Yan-hui, DENG Zi-he, SUN Lin-lin, WU Zhan-sheng. Screening of High-efficiency Cellulose-degrading Microorganism from Spent Lentinula edodes Substrate and Optimization of Its Enzyme Production [J]. Biotechnology Bulletin, 2021, 37(11): 158-165. |
| [9] | CHEN Yi-dan, ZHANG Yu, YANG Jie, ZHANG Qin, JIANG Li. Exploration of Key Functional Genes Affecting Milk Production Traits in Dairy Cattle Based on RNA-seq [J]. Biotechnology Bulletin, 2020, 36(9): 244-252. |
| [10] | WANG Jing-yu, WANG Meng, PAN Yang-yang, WANG Jing-lei, ZHANG Rui, MA Rui, HU Xue-quan, QIU Xiao-fei, CUI Yan, YU Si-jiu, XU Geng-quan. Molecular Characteristics of Bosgrunniens FAF1 Gene and Its Expression in Ovaries,Fallopian Tubes and Uterus at Different Stages [J]. Biotechnology Bulletin, 2020, 36(7): 80-89. |
| [11] | WU Jia-jin, ZHU Sen-lin, ZHOU Mi, SUN Hui-zeng. Research Progress and Trends on Rumen Microbiota in Dairy Cows [J]. Biotechnology Bulletin, 2020, 36(2): 27-38. |
| [12] | YANG Rui-jia, ZHANG Zhong-bao, WU Zhong-yi. Progress of the Structural and Functional Analysis of Plant Transcription Factor TIFY Protein Family [J]. Biotechnology Bulletin, 2020, 36(12): 121-128. |
| [13] | WU Yi, MA Hong-fei, CAO Yong-jia, SI Jing, CUI Bao-kai. Advances on Properties,Production,Purification and Immobilization of Fungal Laccase [J]. Biotechnology Bulletin, 2019, 35(9): 1-10. |
| [14] | QIU Shi-zheng, LI Jia-yi, YANG Jing-chen, LIU Chang-li. Research Progress of Low-cost Method of Synthetizing Polyhydroxyalkanoates(PHAs) [J]. Biotechnology Bulletin, 2019, 35(9): 45-52. |
| [15] | LIU Zhao-shu, LIU Yuan-yuan, QIN Yu-jie, BI Quan, WANG Sai-sai, ZHU Jian-bo. Cloning and Expression Analysis of SikCML7 Gene from Saussurea involucrata [J]. Biotechnology Bulletin, 2019, 35(6): 48-54. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||