








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (11): 162-174.doi: 10.13560/j.cnki.biotech.bull.1985.2022-0321
Previous Articles Next Articles
CAO Ying-hui1( ), HU Mei-juan1, TONG Yan1, ZHANG Yan-ping1, ZHAO Kai2, PENG Dong-hui1, ZHOU Yu-zhen1(
), HU Mei-juan1, TONG Yan1, ZHANG Yan-ping1, ZHAO Kai2, PENG Dong-hui1, ZHOU Yu-zhen1( )
)
Received:2022-03-14
Online:2022-11-26
Published:2022-12-01
Contact:
ZHOU Yu-zhen
E-mail:cyhfred0078@163.com;zhouyuzhencn@163.com
CAO Ying-hui, HU Mei-juan, TONG Yan, ZHANG Yan-ping, ZHAO Kai, PENG Dong-hui, ZHOU Yu-zhen. Identification of the ABC Gene Family and Expression Pattern Analysis During Flower Development in Cymbidium ensifolium[J]. Biotechnology Bulletin, 2022, 38(11): 162-174.
| 基因名称Gene name | 上游引物Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|
| CeABCB6 | GGAGCTACCCTTGTTCGAGA | TGGACTCTGAGCCTTGTGAG |
| CeABCB30 | AGGCCCTTCGTAACCTTTCA | CAAACTTACGCACGACCACA |
| CeABCG3 | AGGGAAAGAGCAGCAGGAAT | TGGCAGCTGTCCATTCAAAC |
| CeABCG54 | ATCAAGCACACAAGGCCAAC | AAGGTGAGGGAAAGTGGCAT |
| CeABCI7 | TGGGGAAGGATATCAGCGAC | CCATGTGCTTGCCTCCAAAT |
| TUB | GCAGTTTACGGCGATGTTCA | ACTCTTCCTCGTCAGCTGTG |
Table 1 Primers used for quantitative real-time PCR
| 基因名称Gene name | 上游引物Forward primer(5'-3') | 下游引物 Reverse primer(5'-3') |
|---|---|---|
| CeABCB6 | GGAGCTACCCTTGTTCGAGA | TGGACTCTGAGCCTTGTGAG |
| CeABCB30 | AGGCCCTTCGTAACCTTTCA | CAAACTTACGCACGACCACA |
| CeABCG3 | AGGGAAAGAGCAGCAGGAAT | TGGCAGCTGTCCATTCAAAC |
| CeABCG54 | ATCAAGCACACAAGGCCAAC | AAGGTGAGGGAAAGTGGCAT |
| CeABCI7 | TGGGGAAGGATATCAGCGAC | CCATGTGCTTGCCTCCAAAT |
| TUB | GCAGTTTACGGCGATGTTCA | ACTCTTCCTCGTCAGCTGTG |
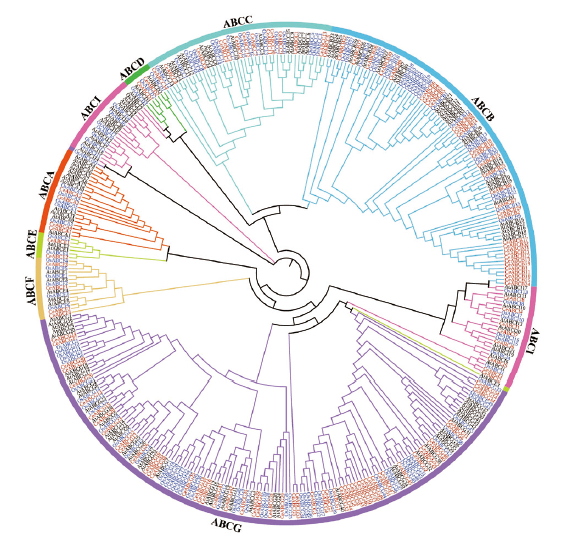
Fig. 4 Phylogenetic analysis of ABC gene family in C. ensifolium,O. sativa and A. thaliana The red font refers to C. ensifolium ABC transporter proteins,the blue font refers to O. sativa ABC transporter proteins,the black font refers to A. thaliana ABC transporter proteins
| 基因名称Gene name | 基因名称Gene name | 复制类型Duplication type | 非同义替换率Ka | 同义替换率Ks | 非同义替换率/同义替换率Ka/Ks |
|---|---|---|---|---|---|
| CeABCG31 | CeABCG29 | 片段复制 | 0.148 156 999 | 0.608 282 47 | 0.243 566 117 |
| CeABCG19 | CeABCG25 | 片段复制 | 0.368 169 066 | NaN | NaN |
| CeABCB18 | CeABCB16 | 片段复制 | 0.069 986 027 | 0.674 387 106 | 0.103 777 231 |
| CeABCG10 | CeABCG46 | 串联复制 | 0.017 153 167 | 0.023 289 543 | 0.736 517 994 |
| CeABCG5 | CeABCG8 | 串联复制 | 0.108 286 131 | 0.156 200 883 | 0.693 249 159 |
| CeABCG8 | CeABCG50 | 串联复制 | 0.016 788 187 | 0.022 178 207 | 0.756 967 701 |
| CeABCG47 | CeABCG1 | 串联复制 | 0.207 255 225 | 2.066 524 894 | 0.100 291 666 |
| CeABCC4 | CeABCC3 | 串联复制 | 0.212 818 781 | 1.459 333 628 | 0.145 832 849 |
| CeABCG18 | CeABCG21 | 串联复制 | 0.006 383 502 | 0.021 424 752 | 0.297 949 888 |
| CeABCI2 | CeABCI3 | 串联复制 | 0.005 878 541 | NaN | NaN |
| CeABCC1 | CeABCC5 | 串联复制 | 0.291 628 981 | NaN | NaN |
| CeABCB12 | CeABCB21 | 串联复制 | 0.024 644 867 | 0.050 781 874 | 0.485 308 334 |
| CeABCB17 | CeABCB16 | 串联复制 | 0.644 437 783 | NaN | NaN |
| CeABCB10 | CeABCB31 | 串联复制 | 0.039 511 484 | 0.03 561 5 | 1.109 405 7 |
| CeABCG30 | CeABCG39 | 串联复制 | 0.106 510 279 | 0.544 293 256 | 0.195 685 466 |
| CeABCG24 | CeABCG27 | 串联复制 | 0.040 625 407 | 0.058 053 477 | 0.699 792 83 |
| CeABCG32 | CeABCG44 | 串联复制 | 0.064 248 894 | 0.099 652 01 | 0.644 732 544 |
Table 2 Nonsynonymous and synonymous substitution ratio calculation of ABC hologous genes in C. ensifolium
| 基因名称Gene name | 基因名称Gene name | 复制类型Duplication type | 非同义替换率Ka | 同义替换率Ks | 非同义替换率/同义替换率Ka/Ks |
|---|---|---|---|---|---|
| CeABCG31 | CeABCG29 | 片段复制 | 0.148 156 999 | 0.608 282 47 | 0.243 566 117 |
| CeABCG19 | CeABCG25 | 片段复制 | 0.368 169 066 | NaN | NaN |
| CeABCB18 | CeABCB16 | 片段复制 | 0.069 986 027 | 0.674 387 106 | 0.103 777 231 |
| CeABCG10 | CeABCG46 | 串联复制 | 0.017 153 167 | 0.023 289 543 | 0.736 517 994 |
| CeABCG5 | CeABCG8 | 串联复制 | 0.108 286 131 | 0.156 200 883 | 0.693 249 159 |
| CeABCG8 | CeABCG50 | 串联复制 | 0.016 788 187 | 0.022 178 207 | 0.756 967 701 |
| CeABCG47 | CeABCG1 | 串联复制 | 0.207 255 225 | 2.066 524 894 | 0.100 291 666 |
| CeABCC4 | CeABCC3 | 串联复制 | 0.212 818 781 | 1.459 333 628 | 0.145 832 849 |
| CeABCG18 | CeABCG21 | 串联复制 | 0.006 383 502 | 0.021 424 752 | 0.297 949 888 |
| CeABCI2 | CeABCI3 | 串联复制 | 0.005 878 541 | NaN | NaN |
| CeABCC1 | CeABCC5 | 串联复制 | 0.291 628 981 | NaN | NaN |
| CeABCB12 | CeABCB21 | 串联复制 | 0.024 644 867 | 0.050 781 874 | 0.485 308 334 |
| CeABCB17 | CeABCB16 | 串联复制 | 0.644 437 783 | NaN | NaN |
| CeABCB10 | CeABCB31 | 串联复制 | 0.039 511 484 | 0.03 561 5 | 1.109 405 7 |
| CeABCG30 | CeABCG39 | 串联复制 | 0.106 510 279 | 0.544 293 256 | 0.195 685 466 |
| CeABCG24 | CeABCG27 | 串联复制 | 0.040 625 407 | 0.058 053 477 | 0.699 792 83 |
| CeABCG32 | CeABCG44 | 串联复制 | 0.064 248 894 | 0.099 652 01 | 0.644 732 544 |

Fig. 6 Collinearity analysis of ABC genes between C. ensifolium and A. thaliana,O. sativa,P. aphrodite Red line indicates the collinearity of pair of ABC genes

Fig. 8 Expression profiles of ABC genes of C. ensifolium in flower development A:Expression patterns related to ABC subfamilies. B:Hierarchical clusters based on gene expression. ZL:Budding flower. CK:Half flowering stage. SK:Full bloomed stage. SB:Senescence stage
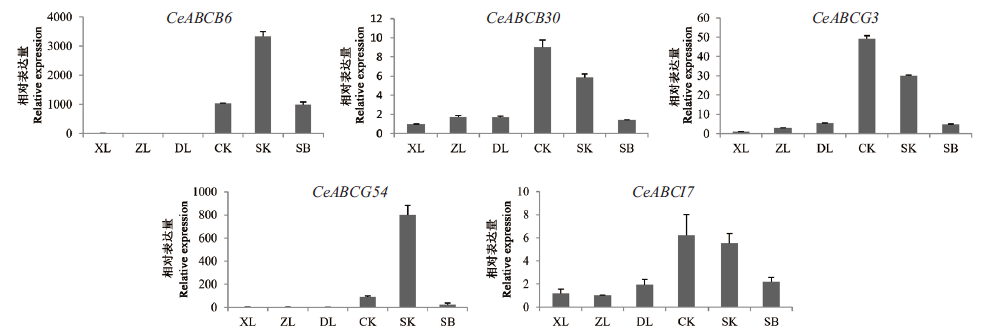
Fig. 9 Express analysis of CeABC genes during flowering development XL:Small bud stage. ZL:Medium bud stage. DL:Big bud stage. CK:Half flowering stage. SK:Full bloomed stage. SB:Senescence stage
| [1] |
Theodoulou FL, Kerr ID. ABC transporter research:going strong 40 years on[J]. Biochem Soc Trans, 2015, 43(5):1033-1040.
doi: 10.1042/BST20150139 URL |
| [2] |
Garcia O, Bouige P, Forestier C, et al. Inventory and comparative analysis of rice and Arabidopsis ATP-binding cassette(ABC)systems[J]. J Mol Biol, 2004, 343(1):249-265.
doi: 10.1016/j.jmb.2004.07.093 URL |
| [3] |
Andolfo G, Ruocco M, di Donato A, et al. Genetic variability and evolutionary diversification of membrane ABC transporters in plants[J]. BMC Plant Biol, 2015, 15:51.
doi: 10.1186/s12870-014-0323-2 pmid: 25850033 |
| [4] |
Do THT, et al. Functions of ABC transporters in plant growth and development[J]. Curr Opin Plant Biol, 2018, 41:32-38.
doi: S1369-5266(17)30151-6 pmid: 28854397 |
| [5] |
Hwang JU, Song WY, Hong D, et al. Plant ABC transporters enable many unique aspects of a terrestrial plant’s lifestyle[J]. Mol Plant, 2016, 9(3):338-355.
doi: 10.1016/j.molp.2016.02.003 URL |
| [6] | Zhang ZL, Tong T, Fang YX, et al. Genome-wide identification of barley ABC genes and their expression in response to abiotic stress treatment[J]. Plants(Basel), 2020, 9(10):1281. |
| [7] |
Chen PJ, Li Y, Zhao LH, et al. Genome-wide identification and expression profiling of ATP-binding cassette(ABC)transporter gene family in pineapple(Ananas comosus(L.)merr. )reveal the role of AcABCG38 in pollen development[J]. Front Plant Sci, 2017, 8:2150.
doi: 10.3389/fpls.2017.02150 URL |
| [8] |
Pang KY, Li YJ, Liu MH, et al. Inventory and general analysis of the ATP-binding cassette(ABC)gene superfamily in maize(Zea mays L.)[J]. Gene, 2013, 526(2):411-428.
doi: 10.1016/j.gene.2013.05.051 URL |
| [9] |
Gani U, Vishwakarma RA, Misra P. Membrane transporters:the key drivers of transport of secondary metabolites in plants[J]. Plant Cell Rep, 2021, 40(1):1-18.
doi: 10.1007/s00299-020-02599-9 URL |
| [10] |
Borghi L, Kang J, de Brito Francisco R. Filling the gap:functional clustering of ABC proteins for the investigation of hormonal transport in planta[J]. Front Plant Sci, 2019, 10:422.
doi: 10.3389/fpls.2019.00422 URL |
| [11] |
Ai Y, et al. The Cymbidium genome reveals the evolution of unique morphological traits[J]. Hortic Res, 2021, 8(1):255.
doi: 10.1038/s41438-021-00683-z URL |
| [12] |
Verrier PJ, et al. Plant ABC proteins—a unified nomenclature and updated inventory[J]. Trends Plant Sci, 2008, 13(4):151-159.
doi: 10.1016/j.tplants.2008.02.001 pmid: 18299247 |
| [13] |
Lefèvre F, Boutry M. Towards identification of the substrates of ATP-binding cassette transporters[J]. Plant Physiol, 2018, 178(1):18-39.
doi: 10.1104/pp.18.00325 pmid: 29987003 |
| [14] |
Sánchez-Fernández R, Davies TG, Coleman JO, et al. The Arabidopsis thaliana ABC protein superfamily, a complete inventory[J]. J Biol Chem, 2001, 276(32):30231-30244.
doi: 10.1074/jbc.M103104200 pmid: 11346655 |
| [15] | Lopez-Ortiz C, Dutta SK, et al. Genome-wide identification and gene expression pattern of ABC transporter gene family in Capsicum spp[J]. PLoS One, 2019, 14(4):e0215901. |
| [16] | Ofori PA, Mizuno A, Suzuki M, et al. Genome-wide analysis of ATP binding cassette(ABC)transporters in tomato[J]. PLoS One, 2018, 13(7):e0200854. |
| [17] |
Zhang XD, Zhao KX, Yang ZM. Identification of genomic ATP binding cassette(ABC)transporter genes and Cd-responsive ABCs in Brassica napus[J]. Gene, 2018, 664:139-151.
doi: S0378-1119(18)30434-7 pmid: 29709635 |
| [18] |
Huang JJ, Li XY, Chen X, et al. Genome-wide identification of soybean ABC transporters relate to aluminum toxicity[J]. Int J Mol Sci, 2021, 22(12):6556.
doi: 10.3390/ijms22126556 URL |
| [19] |
Yan L, Zhang JH, Chen HY, et al. Genome-wide analysis of ATP-binding cassette transporter provides insight to genes related to bioactive metabolite transportation in Salvia miltiorrhiza[J]. BMC Genomics, 2021, 22(1):315.
doi: 10.1186/s12864-021-07623-0 pmid: 33933003 |
| [20] |
Khan N, You FM, Datla R, et al. Genome-wide identification of ATP binding cassette(ABC)transporter and heavy metal associated(HMA)gene families in flax(Linum usitatissimum L.)[J]. BMC Genomics, 2020, 21(1):722.
doi: 10.1186/s12864-020-07121-9 URL |
| [21] | Yan C, et al. Genome-wide identification, evolution, and expression analysis of the ATP-binding cassette transporter gene family in Brassica rapa[J]. Front Plant Sci, 2017, 8:349. |
| [22] |
Cui BL, Li Y, Zhang YJ. A comparative genome-wide analysis of the ABC transporter gene family among three Gossypium species[J]. Crop Sci, 2021, 61(4):2489-2509.
doi: 10.1002/csc2.20525 URL |
| [23] |
Lane TS, Rempe CS, Davitt J, et al. Diversity of ABC transporter genes across the plant kingdom and their potential utility in biotechnology[J]. BMC Biotechnol, 2016, 16(1):47.
doi: 10.1186/s12896-016-0277-6 pmid: 27245738 |
| [24] |
Geisler M, Aryal B, di Donato M, et al. A critical view on ABC transporters and their interacting partners in auxin transport[J]. Plant Cell Physiol, 2017, 58(10):1601-1614.
doi: 10.1093/pcp/pcx104 pmid: 29016918 |
| [25] |
Panikashvili D, Shi JX, Schreiber L, et al. The Arabidopsis ABCG13 transporter is required for flower cuticle secretion and patterning of the petal epidermis[J]. New Phytol, 2011, 190(1):113-124.
doi: 10.1111/j.1469-8137.2010.03608.x pmid: 21232060 |
| [26] |
Kuromori T, Miyaji T, et al. ABC transporter AtABCG25 is involved in abscisic acid transport and responses[J]. Proc Natl Acad Sci USA, 2010, 107(5):2361-2366.
doi: 10.1073/pnas.0912516107 URL |
| [27] |
Kang J, Hwang JU, Lee M, et al. PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid[J]. Proc Natl Acad Sci USA, 2010, 107(5):2355-2360.
doi: 10.1073/pnas.0909222107 URL |
| [28] |
Demissie ZA, Tarnowycz M, Adal AM, et al. A lavender ABC transporter confers resistance to monoterpene toxicity in yeast[J]. Planta, 2019, 249(1):139-144.
doi: 10.1007/s00425-018-3064-x pmid: 30535718 |
| [29] |
Adebesin F, Widhalm JR, Boachon B, et al. Emission of volatile organic compounds from Petunia flowers is facilitated by an ABC transporter[J]. Science, 2017, 356(6345):1386-1388.
doi: 10.1126/science.aan0826 pmid: 28663500 |
| [30] | 彭红明. 中国兰花挥发及特征花香成分研究[D]. 北京: 中国林业科学研究院, 2009. |
| Peng HM. Study on the volatile, characteristic floral fragrance components of Chinese Cymbidium[D]. Beijing: Chinese Academy of Forestry, 2009. | |
| [31] |
Huang MK, Ma CP, Yu RC, et al. Concurrent changes in methyl jasmonate emission and the expression of its biosynthesis-related genes in Cymbidium ensifolium flowers[J]. Physiol Plant, 2015, 153(4):503-512.
doi: 10.1111/ppl.12275 URL |
| [32] | 李梦雅. 茉莉酸转运蛋白的发现及其在系统伤害抗性中传递系统信号的功能分析[D]. 北京: 中国农业大学, 2014. |
| Li MY. The identification of jasmonate transporters and their essential role of signal relay in systemic wound resistance. Beijing: China Agricultural University, 2014. |
| [1] | ZHANG Lu-yang, HAN Wen-long, XU Xiao-wen, YAO Jian, LI Fang-fang, TIAN Xiao-yuan, ZHANG Zhi-qiang. Identification and Expression Analysis of the Tobacco TCP Gene Family [J]. Biotechnology Bulletin, 2023, 39(6): 248-258. |
| [2] | LAI Rui-lian, FENG Xin, GAO Min-xia, LU Yu-dan, LIU Xiao-chi, WU Ru-jian, CHEN Yi-ting. Genome-wide Identification of Catalase Family Genes and Expression Analysis in Kiwifruit [J]. Biotechnology Bulletin, 2023, 39(4): 136-147. |
| [3] | ZHAO Yan-xia, ZHANG Jing-ying, SUN Jun-fei, WANG Jiang-hui, SUN Jia-bo, LV Xiao-hui. Analyses of Transcription and Metabolic Differential in the Flower Development Processes of ‘Rose rugosa cv. Plena’ [J]. Biotechnology Bulletin, 2023, 39(3): 184-195. |
| [4] | DUAN Min-jie, LI Yi-fei, YANG Xiao-miao, WANG Chun-ping, HUANG Qi-zhong, HUANG Ren-zhong, ZHANG Shi-cai. Identification of Zinc Finger Protein DnaJ-Like Gene Family in Capsicum annuum and Its Expression Analysis Responses to High Temperature Stress [J]. Biotechnology Bulletin, 2023, 39(1): 187-198. |
| [5] | YUAN Xing, GUO Cai-hua, LIU Jin-ming, KANG Chao, QUAN Shao-wen, NIU Jian-xin. Genome-wide Identification of CONSTANS-Like Family Genes and Expression Analysis in Wanlut [J]. Biotechnology Bulletin, 2022, 38(9): 167-179. |
| [6] | GUO Bin-hui, SONG Li. Transcription of Ethylene Biosynthesis and Signaling Associated Genes in Response to Heterodera glycine Infection [J]. Biotechnology Bulletin, 2022, 38(8): 150-158. |
| [7] | GAO Ling, WANG Fei, XIE Shuang-quan, CHEN Xi-feng, SHEN Hai-tao, LI Hong-bin. Genome-wide Identification and Expression Analysis of CBL Gene Family in Glycyrrhiza uralensis [J]. Biotechnology Bulletin, 2021, 37(4): 18-27. |
| [8] | YANG Yi-hong, XU Hao, CHEN Duan-fen, GAO Zhi-min. Research Advances on the Gene for Gibberellin 3-Beta-Dioxygenase in Higher Plants [J]. Biotechnology Bulletin, 2018, 34(3): 18-22. |
| [9] | Yan Jun, Guo Xingqi, Cao Xuecheng. Progress of Genome-wide Researches on WRKY Transcription Factors [J]. Biotechnology Bulletin, 2015, 31(11): 9-17. |
| [10] | Jiang Wei, Gu Huiying, Wang Zhimin, Song Ming, Tang Qinglin. The Flower Development of Arabidopsis thaliana Affected by Floral Meristem Identity Gene AGL24 [J]. Biotechnology Bulletin, 2014, 0(4): 6-13. |
| [11] | Li Jing, Gu Huiying, Wang Zhimin, Tang Qinglin, Song Ming. Research Progress of Flowering Gene Regulatory Networks in Arabidopsis thaliana [J]. Biotechnology Bulletin, 2014, 0(12): 1-8. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||