








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (7): 269-277.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1299
Previous Articles Next Articles
WANG Guang-li1,2( ), FAN Chan1,2, WANG Hui1,2, LU Hui-fang1,2, XIA Ling-yin1,2, HUANG Jian1,2, MIN Xun1,2(
), FAN Chan1,2, WANG Hui1,2, LU Hui-fang1,2, XIA Ling-yin1,2, HUANG Jian1,2, MIN Xun1,2( )
)
Received:2021-10-14
Online:2022-07-26
Published:2022-08-09
Contact:
MIN Xun
E-mail:1947952722@qq.com;zmchj2001@163.com
WANG Guang-li, FAN Chan, WANG Hui, LU Hui-fang, XIA Ling-yin, HUANG Jian, MIN Xun. Prokaryotic Expression,Purification,Identification,and Polyclonal Antibody Preparation of Vibrio cholerae Hemolysin HlyA[J]. Biotechnology Bulletin, 2022, 38(7): 269-277.
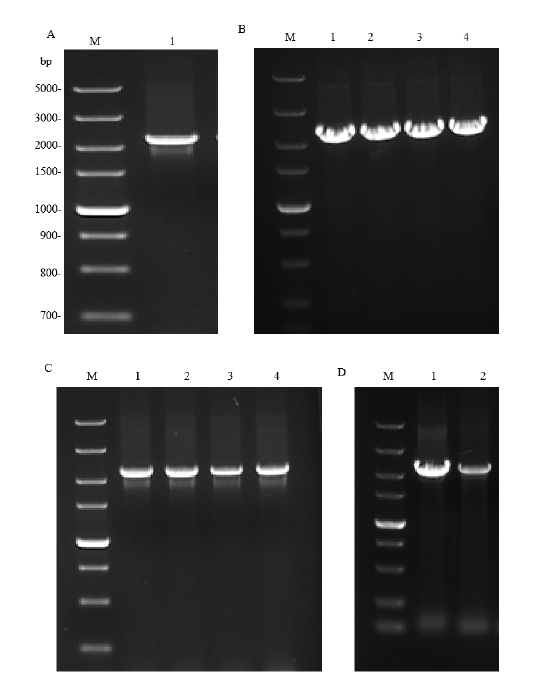
Fig.1 Construction of recombinant plasmids pET28a-hlyA,pET32 a-hlyA and pCold TF-hlyA. M:DNA marker DL5000 bp;A:electrophoresis results of hlyA gene PCR amplification(1:hlyA fragment). B-D:Electrophoresis results of pET28a-hlyA,pET32a-hlyA and pCold TF-hlyArecombinant plasmid bacterial liquid PCR,respectively(1:positive control;2-4:positive cloning bacteria)
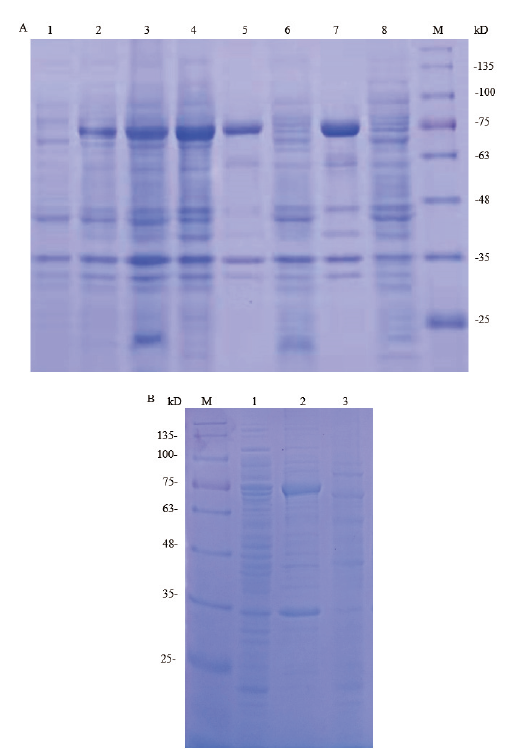
Fig.2 SDS-PAGE analysis of pET28a-hlyA expression pro-ducts in E. coli BL21(DE3)induced with IPTG M:180 kD protein marker;A:1:uninduced E. coli BL21(DE3)with recombinant plasmid pET28a-hlyA;2-3:induced E. coli BL21(DE3)with recombinant plasmid pET28a-hlyA at 10℃,120 r/min by 0.05 mmol/L and 0.1 mmol/L IPTG for 16 h;4:induced E. coli BL21(DE3)with recombinant plasmid pET28a-hlyA at 15℃,100 r/min by 0.05 mmol/L IPTG for 12 h;5 and 7:lysate pellet of induced bacteria;6 and 8:lysate supernatant of induced bacteria. B:1:uninduced E. coli BL21(DE3)with recombinant plasmid pET28a-hlyA;2:lysate pellet of induced bacteria at 23℃ by 0.05 mmol/L IPTG for 10 h;3:lysate supernatant of induced bacteria at 23℃
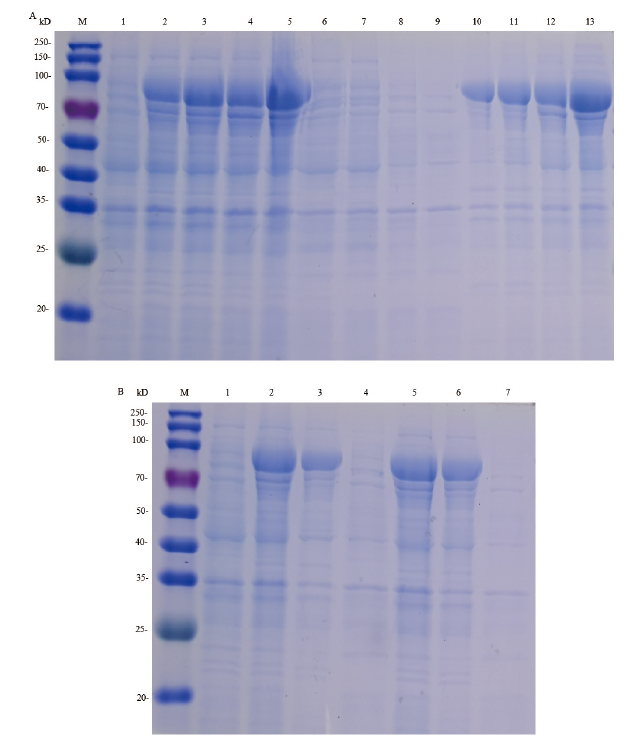
Fig.3 SDS-PAGE analysis of pET32a-hlyA expression pr-oducts in E. coli BL21(DE3)induced with IPTG M:250 kD protein marker. A:1:uninduced E. coli BL21(DE3)with recombinant plasmid pET32a-hlyA;2-3:induced E. coli BL21(DE3)with recombinant plasmid pET32a-hlyA at 15℃,100 r/min by 0.05 and 0.1 mmol/L IPTG for 16 h;4 and 5:induced E. coli BL21(DE3)with recombinant plasmid pET32a-hlyA at 25℃,120 r/min by 0.05 and 0.1 mmol/L IPTG for 10 h;6-9:lysate supernatant of induced bacteria;10-13:lysate pellet of induced bacteria. B:1:uninduced E. coli BL21(DE3)with recombinant plasmid pET32a-hlyA;2 and 5:induced E. coliBL21(DE3)with recombinant plasmid pET32a-hlyA at 20℃,100 r/min by 0.05 and 0.1 mmol/L IPTG for 12 h;3 and 6:lysate pellet of induced bacteria;4 and 7:lysate supernatant of induced bacteria

Fig.4 SDS-PAGE analysis of pCold TF-hlyA expression products in E. coli BL21 induced with IPTG and the purified HlyA protein M:250 kD protein marker;1:uninduced E. coli BL21 with recombinant plasmid pCold TF -hlyA;2:induced E. coli BL21 with recombinant plasmid pCold TF -hlyA;3:lysate supernatant of induced bacteria;4-9:1×Washing Buffer contain of 10,20,30,40,300,500 mmol/L imidazole through Ni-chelating affinity chromatography
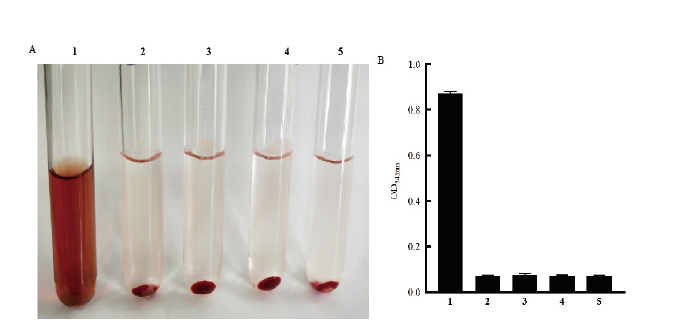
Fig.5 Hemolytic activity determination of recombinant expressive protein HlyA 1:Positive control(1% Trion-X);2:negative control(PBS);3:0 μg HlyA protein incubateed with 2% rabbit red blood cells;4:4 μg HlyA protein incubateed with 2% rabbit red blood cells;5:8 μg HlyA protein co-incubated with 2% rabbit red blood cells
Fig.8 Mass spectrometric profiles of HlyA protein A mass spectrum of the peptide sequence SASFTVDWDHPVFTGGRPVNLQLAS-FNNR that matches the amino acid sequence of the HlyA protein is shown
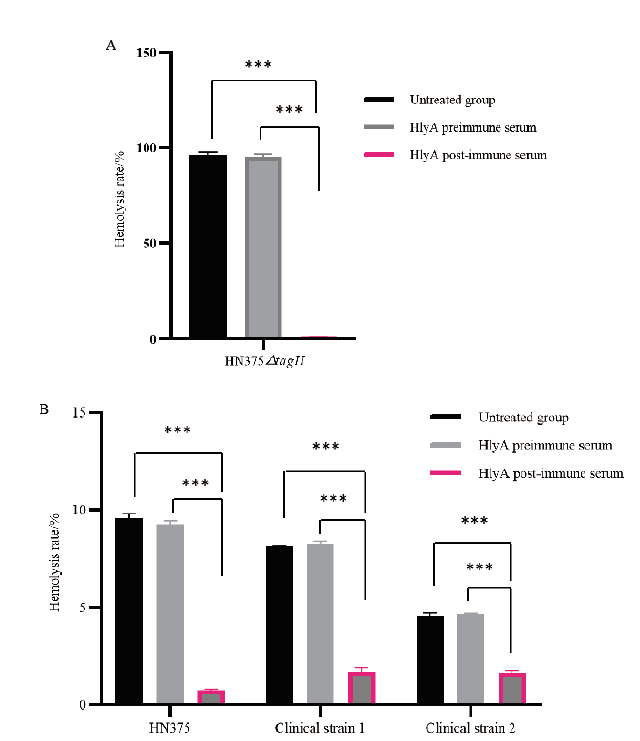
Fig. 9 Detection of Anti-HlyA polyclonal antibody neutrali-zation activity A:Analysis of the inhibitory effect of anti-HlyA polyclonal antibody on the hemolytic activity of HN375△tagH strain;B:Analysis of the inhibitory effect of anti-HlyA polyclonal antibody on the hemolytic activity of HN375 strain and two clinical strains. *** indicates that there is a significant difference in the data between two groups(P < 0.001)
| [1] |
Saka HA, Bidinost C, Sola C, et al. Vibrio cholerae cytolysin is essential for high enterotoxicity and apoptosis induction produced by a cholera toxin gene-negative V. cholerae non-O1, non-O139 strain[J]. Microb Pathog, 2008. 44(2):118-28.
doi: 10.1016/j.micpath.2007.08.013 URL |
| [2] | Ruenchit P, Reamtong O, Siripanichgon K, Chaicumpa W, Diraphat P. New facet of non-O1/non-O139 Vibrio cholerae hemolysin A:a competitive factor in the ecological niche[J]. FEMS Microbiol Ecol, 2017. 93(12). |
| [3] |
Zmeter C, Tabaja H, Sharara AI, Kanj SS. Non-O1, non-O139 Vibrio cholerae septicemia at a tertiary care center in Beirut, Lebanon;a case report and review[J]. J Infect Public Health, 2018. 11(5):601-604.
doi: 10.1016/j.jiph.2018.01.001 URL |
| [4] |
Jiang F, Bi R, Deng L, Kang H, Gu B, Ma P. Virulence-associated genes and molecular characteristics of non-O1/non-O139 Vibrio cholerae isolated from hepatitis B cirrhosis patients in China[J]. Int J Infect Dis, 2018. 74:117-122.
doi: S1201-9712(18)34456-4 pmid: 29969728 |
| [5] |
Dobrović K, Rudman F, Ottaviani D, Šestan Crnek S, Leoni F, Škrlin J. A rare case of necrotizing fasciitis caused by Vibrio cholerae O8 in an immunocompetent patient[J]. Wien Klin Wochenschr, 2016. 128(19-20):728-730.
doi: 10.1007/s00508-016-1060-3 URL |
| [6] |
Hao Y, Wang Y, Bi Z, et al. A case of non-O1/non-O139 Vibrio cholerae septicemia and meningitis in a neonate[J]. Int J Infect Dis, 2015. 35:117-9.
doi: 10.1016/j.ijid.2015.05.004 URL |
| [7] | Marinello S, Marini G, Parisi G, et al. Vibrio cholerae non-O1, non-O139 bacteraemia associated with pneumonia[J]. Italy 2016. Infection, 2017. 45(2):237-240. |
| [8] |
Fan Y, Li Z, Li Z, et al. Nonhemolysis of epidemic El Tor biotype strains of Vibrio cholerae is related to multiple functional deficiencies of hemolysin A[J]. Gut Pathog, 2019. 11:38.
doi: 10.1186/s13099-019-0316-7 URL |
| [9] | Chen YT, Tang HJ, Chao CM, Lai CC. Clinical manifestations of non-O1 Vibrio cholerae infections[J]. PLoS One,2015. 10(1):e0116904. |
| [10] | Chen J, Huang J, Huang M, et al. Two cases of septic shock with different outcomes caused by non-O1/non-O139 Vibrio cholerae isolates[J]. J Int Med Res,2020. 48(6):300060520933459. |
| [11] | Chatterjee S, Ghosh K, Raychoudhuri A, et al. Incidence, virulence factors, and clonality among clinical strains of non-O1, non-O139 Vibrio cholerae isolates from hospitalized diarrheal patients in Kolkata[J]. India. J Clin Microbiol, 2009. 47(4):1087-95. |
| [12] |
Mukherjee A, Ganguly S, Chatterjee NS, Banerjee KK. Vibrio cholerae hemolysin:The β-trefoil domain is required for folding to the native conformation[J]. Biochem Biophys Rep, 2016. 8:242-248.
doi: 10.1016/j.bbrep.2016.09.009 pmid: 28955962 |
| [13] |
Ganguly S, Mukherjee A, Mazumdar B, Ghosh AN, Banerjee KK. The β-prism lectin domain of Vibrio cholerae hemolysin promotes self-assembly of the β-pore-forming toxin by a carbohydrate-independent mechanism[J]. J Biol Chem,2014. 289(7):4001-8.
doi: 10.1074/jbc.M113.522284 URL |
| [14] |
Diep TT, Nguyen NT, Nguyen TN, et al. Isolation of New Delhi metallo-β-lactamase 1-producing Vibrio cholerae non-O1, non-O139 strain carrying ctxA, st and hly genes in southern Vietnam[J]. Microbiol Immunol, 2015. 59(5):262-7.
doi: 10.1111/1348-0421.12248 URL |
| [15] |
Saha N, Banerjee KK. Carbohydrate-mediated regulation of interaction of Vibrio cholerae hemolysin with erythrocyte and phospholipid vesicle[J]. J Biol Chem, 1997. 272(1):162-7.
doi: 10.1074/jbc.272.1.162 URL |
| [16] |
Yamamoto K, Al-Omani M, Honda T, Takeda Y, Miwatani T. Non-O1 Vibrio cholerae hemolysin:purification, partial characterization, and immunological relatedness to El Tor hemolysin[J]. Infect Immun,1984. 45(1):192-6.
doi: 10.1128/iai.45.1.192-196.1984 URL |
| [17] |
Coelho A, Andrade JR, Vicente AC, Dirita VJ. Cytotoxic cell vacuolating activity from Vibrio cholerae hemolysin[J]. Infect Immun,2000. 68(3):1700-5.
doi: 10.1128/IAI.68.3.1700-1705.2000 URL |
| [18] |
Figueroa-Arredondo P, Heuser JE, Akopyants NS, et al. Cell vacuolation caused by Vibrio cholerae hemolysin[J]. Infect Immun, 2001. 69(3):1613-24.
doi: 10.1128/IAI.69.3.1613-1624.2001 URL |
| [19] |
Chakraborty DC, Mukherjee G, Banerjee P, Banerjee KK, Biswas T. Hemolysin induces Toll-like receptor(TLR)-independent apoptosis and multiple TLR-associated parallel activation of macrophages[J]. J Biol Chem,2011. 286(40):34542-51.
doi: 10.1074/jbc.M111.241851 URL |
| [20] | Debellis L, Diana A, Arcidiacono D, et al. The Vibrio cholerae cytolysin promotes chloride secretion from intact human intestinal mucosa[J]. PLoS One,2009. 4(3):e5074. |
| [21] |
Alm RA, Mayrhofer G, Kotlarski I, Manning PA. Amino-terminal domain of the El Tor haemolysin of Vibrio cholerae O1 is expressed in classical strains and is cytotoxic[J]. Vaccine,1991. 9(8):588-94.
doi: 10.1016/0264-410X(91)90247-4 URL |
| [22] |
Tsou AM, Zhu J. Quorum sensing negatively regulates hemolysin transcriptionally and posttranslationally in Vibrio cholerae[J]. Infect Immun,2010. 78(1):461-7.
doi: 10.1128/IAI.00590-09 URL |
| [23] |
Tsou AM, Cai T, Liu Z, Zhu J, Kulkarni RV. Regulatory targets of quorum sensing in Vibrio cholerae:evidence for two distinct HapR-binding motifs[J]. Nucleic Acids Res,2009. 37(8):2747-56.
doi: 10.1093/nar/gkp121 URL |
| [24] | Nguyen AN, Disconzi E, Charrière GM, et al. csrB gene duplication drives the evolution of redundant regulatory pathways controlling expression of the major toxic secreted metalloproteases in vibrio tasmaniensis LGP32[J]. mSphere,2018. 3(6). |
| [25] |
McCardell BA, Kothary MH, Madden JM. Two-step purification and partial characterization of a variant of the Vibrio cholerae non-O1 hemolysin[J]. FEMS Microbiol Lett,1999. 180(2):177-82.
doi: 10.1111/j.1574-6968.1999.tb08793.x URL |
| [26] |
Dutta S, Mazumdar B, Banerjee KK, Ghosh AN. Three-dimensional structure of different functional forms of the Vibrio cholerae hemolysin oligomer:a cryo-electron microscopic study[J]. J Bacteriol, 2010, 192(1):169-78.
doi: 10.1128/JB.00930-09 URL |
| [27] | 尹鑫, 刘澜澜, 贾莹, 等. 鸡氨肽酶N的高效可溶性表达及生物学功能分析[J]. 生物工程学报, 2010, 26(4):470-475. |
| Yin X, Liu L, Jia Y, et al. Expression and biological function analysis of chicken aminopeptidase N[J]. Chinese Journal of Biotechnology,2010. 26(4):470-5. | |
| [28] |
De S, Olson R. Crystal structure of the Vibrio cholerae cytolysin heptamer reveals common features among disparate pore-forming toxins[J]. Proc Natl Acad Sci USA, 2011, 108(18):7385-90.
doi: 10.1073/pnas.1017442108 URL |
| [29] |
Nagamune K, Yamamoto K, Naka A, Matsuyama J, Miwatani T, Honda T. In vitro proteolytic processing and activation of the recombinant precursor of El Tor cytolysin/hemolysin(pro-HlyA)of Vibrio cholerae by soluble hemagglutinin/protease of V. cholerae, trypsin, and other proteases[J]. Infect Immun, 1996. 64(11):4655-8.
doi: 10.1128/iai.64.11.4655-4658.1996 URL |
| [30] |
Nagamune K, Yamamoto K, Honda T. Intramolecular chaperone activity of the pro-region of Vibrio cholerae El Tor cytolysin[J]. J Biol Chem,1997. 272(2):1338-43.
doi: 10.1074/jbc.272.2.1338 URL |
| [1] | MEI Huan, LI Yue, LIU Ke-meng, LIU Ji-hua. Study on the Biosynthesis of l-SLR by Efficient Prokaryotic Expression of Berberine Bridge Enzyme [J]. Biotechnology Bulletin, 2023, 39(7): 277-287. |
| [2] | TENG Meng-xin, XU Ya, HE Jing, WANG Qi, QIAO Fei, LI Jing-yang, LI Xin-guo. Cloning and Prokaryotic Expression Analysis of MaMC6 in Banana [J]. Biotechnology Bulletin, 2023, 39(12): 179-186. |
| [3] | HOU Wei-chen, YE Ke, LI Jie, ZHANG Yang-zi, XU Wen-tao, ZHU Long-jiao, LI Xiang-yang. Detection of Escherichia coli O157: H7 Based on Antibody Aptamer Sandwich Biosensor [J]. Biotechnology Bulletin, 2023, 39(12): 81-89. |
| [4] | GUO Wen-bo, LU Yang, SUI Li, ZHAO Yu, ZOU Xiao-wei, ZHANG Zheng-kun, LI Qi-yun. Preparation and Application of Polyclonal Antibodies Against Beauveria bassiana Mycovirus BbPmV-4 Coat Protein [J]. Biotechnology Bulletin, 2023, 39(10): 58-67. |
| [5] | SUO Qing-qing, WU Nan, YANG Hui, LI Li, WANG Xi-feng. Prokaryotic Expression,Antibody Preparation and Application of Rice Caffeoyl Coenzyme A-O-methyltransferase Gene [J]. Biotechnology Bulletin, 2022, 38(8): 135-141. |
| [6] | QIN Xue-jing, WANG Yu-han, CAO Yi-bo, ZHANG Ling-yun. Prokaryotic Expression and Preparation of Polyclonal Antibody of PwHAP5 Gene in Picea wilsonii [J]. Biotechnology Bulletin, 2022, 38(8): 142-149. |
| [7] | WANG Qiao-ju, HU Yu-meng, WEN Ya-ya, SONG Li, MENG Chuang, PAN Zhi-ming, JIAO Xin-an. Expression and Activity Identification of SARS-CoV-2 S1 Protein [J]. Biotechnology Bulletin, 2022, 38(3): 157-163. |
| [8] | SHEN Jun-qiang, ZHANG Li-ping, YU Rui-ming, WANG Yong-lu, PAN Li, LIU Xia, LIU Xin-sheng. Porcine Kobuvirus Structural Proteins VP0 and VP1 Prokaryotic Expression and Establishment of Indirect ELISA Method [J]. Biotechnology Bulletin, 2022, 38(10): 243-253. |
| [9] | SHAN Cao-mei, YE Lei, ZHANG Lian-hu, KUANG Wei-gang, SUN Xiao-tang, MA Jian, CUI Ru-qiang. Cloning,and Functional Analysis of Gene OsRAI1 Resistant to Hirschmanniella mucronate in Rice [J]. Biotechnology Bulletin, 2021, 37(7): 146-155. |
| [10] | ZENG Fu-yuan, SU Ze-hui, ZHOU Shi-hui, XIE Miao, PANG Huan-ying. Prokaryotic Expression of the PEPCK Protein of Vibrio alginolyticus and Identification of Its Acetylation and Succinylation [J]. Biotechnology Bulletin, 2021, 37(5): 84-91. |
| [11] | ZHANG Xi-xi, ZHANG Yi-qing, LI Yu-lin, HAN Xiao, WANG Guo-qiang, WANG Xiao-jun, WANG Xu-dong, WANG Yun-long. Prokaryotic Expression,Purification and Application of N Protein C-terminal Recombinant Protein in Novel Coronavirus(SARS-CoV-2) [J]. Biotechnology Bulletin, 2021, 37(5): 92-97. |
| [12] | BAI Fu-mei, LI Zhi-min, WANG Xiao-qin, HU Zi-wei, BAO Ling-ling, LI Zhi-min. Biochemical Characterization and Structural Analysis of N-acetylornithine Transaminase from Synechocystis sp. PCC6803 [J]. Biotechnology Bulletin, 2021, 37(5): 98-107. |
| [13] | QU Huan, LI Cheng, CHEN Rui, LIAO Yi-jie, CAO San-jie, WEN Yi-ping, YAN Qi-gui, HUANG Xiao-bo. Truncated Expression of the S1-CTD Fragment of Porcine Deltacoronavirus and Establishment of an Indirect ELISA for Detecting Its Antibody [J]. Biotechnology Bulletin, 2021, 37(5): 273-280. |
| [14] | PENG Li-zhong, ZHANG Peng, ZHOU Wen-wen, ZENG Xu-hui, ZHANG Xiao-ning. Preparation and Multi-purpose Validation of Sperm-specific Protein Cabs1 Polyclonal Antibody [J]. Biotechnology Bulletin, 2021, 37(3): 261-270. |
| [15] | HE Yang, YU Qiao-ling, WANG Jun, QIN Chuan-jie, LI Hua-tao. Advances in Prokaryotic Expression Gene of Tilapia [J]. Biotechnology Bulletin, 2021, 37(2): 195-202. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||