








Biotechnology Bulletin ›› 2022, Vol. 38 ›› Issue (9): 207-214.doi: 10.13560/j.cnki.biotech.bull.1985.2021-1450
Previous Articles Next Articles
LI Wen-shuo( ), WANG Lin-song, DU Gui-cai, GUO Qun-qun, ZHANG Ting-ting, YANG Hong LI Rong-gui
), WANG Lin-song, DU Gui-cai, GUO Qun-qun, ZHANG Ting-ting, YANG Hong LI Rong-gui
Received:2021-11-21
Online:2022-09-26
Published:2022-10-11
LI Wen-shuo, WANG Lin-song, DU Gui-cai, GUO Qun-qun, ZHANG Ting-ting, YANG Hong LI Rong-gui. Gene Cloning of an Aldehyde Dehydrogenase from Bursaphelenchus xylophilus and Biochemical Characterization[J]. Biotechnology Bulletin, 2022, 38(9): 207-214.
| 引物名称 Name of primer | 序列Sequence(5'-3') |
|---|---|
| F1 | aaaCATATGggacagctccagagtttggt |
| R1 | aaaGGATCCtcactcgatggagatccgctttc |
Table 1 PCR primers used in this study
| 引物名称 Name of primer | 序列Sequence(5'-3') |
|---|---|
| F1 | aaaCATATGggacagctccagagtttggt |
| R1 | aaaGGATCCtcactcgatggagatccgctttc |

Fig. 3 Expression and purification of the recombinant alde-hyde dehydrogenase M:Protein marker;1:total proteins of E. coli BL21(DE3);2:total proteins of E. coli BL21(DE3)harboring pET-15b-aldh;3:the purified recombinant aldehyde dehydrogenase
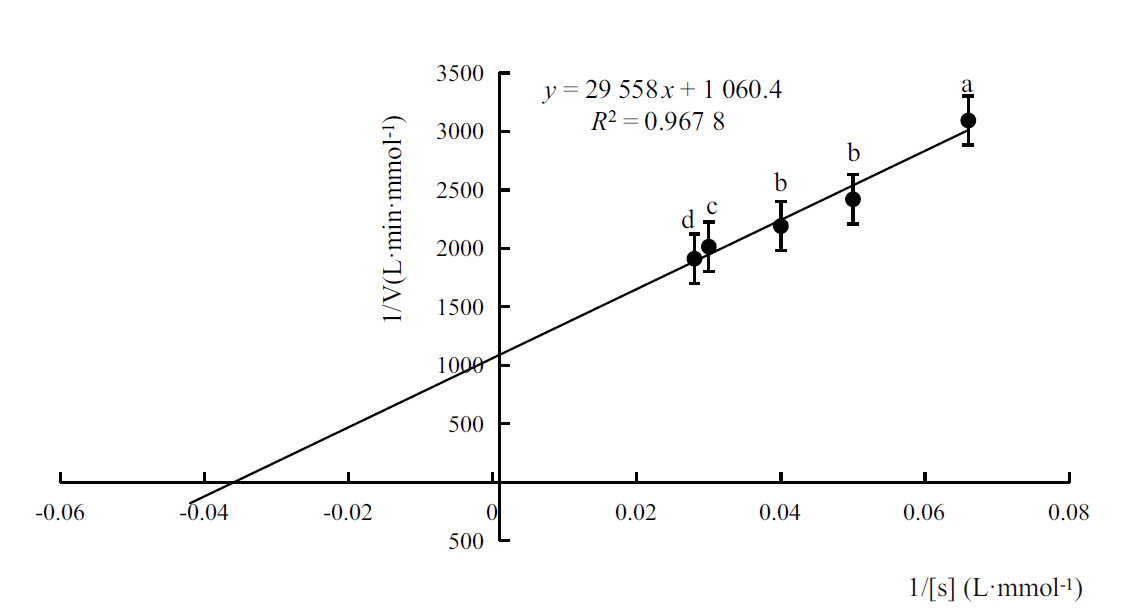
Fig. 4 Km of the recombinant aldehyde dehydrogenase calculated by Lineweaver-Burk plot The error line in the figure refers to the standard deviation. The lowercase letters indicate that the difference among different test reached a significant level(P <0.05). The same meaning is suitable for the rest of this paper

Fig.9 Multiple sequences alignments of primary structures of aldehyde dehydrogenases from different sources Bursaphelenchus xylophilus:GenBank NO.CAD5152421.1;Bursaphelenchus okinawaensis:GenBank NO.CAD5218566.1;Trypanosoma grayi:GenBank NO.XP_009312368.1;Caenorhabditis elegans:GenBank NO. NP_741553.1;Mus musculus:GenBank NO.NP_001318043.1;Strongyloides ratti:GenBank NO. XP_024502805.1;Pseudomonas sp. HR199:GenBank NO. CAA06926.1;Schistocerca gregaria:GenBank NO.QVD39485.1;Meloidogyne graminicola:GenBank NO. KAF7633361.1;Ancylostoma caninum:GenBank NO.RCN51637.1;Micrococcus luteus NCTC 2665:GenBank NO. ACS29670.1
| [1] | Mota MM, Futai K, Vieira P. Pine wilt disease and the pinewood nematode, Bursaphelenchus xylophilus[M]// Ciancio A, Mukerji KG. Integrated Management of Fruit Crops Nematodes. Dordrecht: Springer Netherlands, 2009:253-274. |
| [2] |
Meng FL, Wang J, Wang X, et al. Expression analysis of thaumatin-like proteins from Bursaphelenchus xylophilus and Pinus massoniana[J]. Physiol Mol Plant Pathol, 2017, 100:178-184.
doi: 10.1016/j.pmpp.2017.10.002 URL |
| [3] |
Futai K. Pine wood nematode, Bursaphelenchus xylophilus[J]. Annu Rev Phytopathol, 2013, 51:61-83.
doi: 10.1146/annurev-phyto-081211-172910 URL |
| [4] |
Macho AP, Zipfel C. Plant PRRs and the activation of innate immune signaling[J]. Mol Cell, 2014, 54(2):263-272.
doi: 10.1016/j.molcel.2014.03.028 pmid: 24766890 |
| [5] |
Li Y, Hu LJ, Wu XQ, et al. A Bursaphelenchus xylophilus effector, Bx-FAR-1, suppresses plant defense and affects nematode infection of pine trees[J]. Eur J Plant Pathol, 2020, 157(3):637-650.
doi: 10.1007/s10658-020-02031-8 URL |
| [6] |
Holbein J, Grundler FMW, Siddique S. Plant basal resistance to Nematodes:an update[J]. J Exp Bot, 2016, 67(7):2049-2061.
doi: 10.1093/jxb/erw005 pmid: 26842982 |
| [7] |
Han ZM, Hong YD, Zhao BG. A study on pathogenicity of bacteria carried by pine wood Nematodes[J]. J Phytopathol, 2003, 151(11/12):683-689.
doi: 10.1046/j.1439-0434.2003.00790.x URL |
| [8] |
Zhang W, Yu HY, Lv YX, et al. Gene family expansion of pinewood nematode to detoxify its host defence chemicals[J]. Mol Ecol, 2020, 29(5):940-955.
doi: 10.1111/mec.15378 pmid: 32031723 |
| [9] |
Li Z, Zhang QW, Zhou XG. A 2-Cys peroxiredoxin in response to oxidative stress in the pine wood nematode, Bursaphelenchus xylophilus[J]. Sci Rep, 2016, 6:27438.
doi: 10.1038/srep27438 URL |
| [10] |
Fu HY, Ren JH, Huang L, et al. Screening and functional analysis of the peroxiredoxin specifically expressed in Bursaphelenchus xylophilus-the causative agent of pine wilt disease[J]. Int J Mol Sci, 2014, 15(6):10215-10232.
doi: 10.3390/ijms150610215 URL |
| [11] |
Xu CJ, Li CYT, Kong ANT. Induction of phase I, II and III drug metabolism/transport by xenobiotics[J]. Arch Pharm Res, 2005, 28(3):249-268.
pmid: 15832810 |
| [12] |
Yan X, Cheng XY, et al. Comparative transcriptomics of two pathogenic pinewood nematodes yields insights into parasitic adaptation to life on pine hosts[J]. Gene, 2012, 505(1):81-90.
doi: 10.1016/j.gene.2012.05.041 pmid: 22705985 |
| [13] |
Guengerich FP. Cytochrome p450 and chemical toxicology[J]. Chem Res Toxicol, 2008, 21(1):70-83.
pmid: 18052394 |
| [14] |
Xu XL, Wu XQ, Ye JR, et al. Molecular characterization and functional analysis of three pathogenesis-related cytochrome P450 genes from Bursaphelenchus xylophilus(Tylenchida:Aphelenchoidoidea)[J]. Int J Mol Sci, 2015, 16(3):5216-5234.
doi: 10.3390/ijms16035216 URL |
| [15] |
Wang LS, Zhang TT, Pan ZS, et al. The alcohol dehydrogenase with a broad range of substrate specificity regulates vitality and reproduction of the plant-parasitic nematode Bursaphelenchus xylophilus[J]. Parasitology, 2019, 146(4):497-505.
doi: 10.1017/S0031182018001695 URL |
| [16] |
Koppaka V, Thompson DC, Chen Y, et al. Aldehyde dehydrogenase inhibitors:a comprehensive review of the pharmacology, mechanism of action, substrate specificity, and clinical application[J]. Pharmacol Rev, 2012, 64(3):520-539.
doi: 10.1124/pr.111.005538 URL |
| [17] |
Guo QQ, Du GC, Qi HT, et al. A nematicidal tannin from Punica granatum L. rind and its physiological effect on pine wood nematode(Bursaphelenchus xylophilus)[J]. Pestic Biochem Physiol, 2017, 135:64-68.
doi: 10.1016/j.pestbp.2016.06.003 URL |
| [18] |
Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding[J]. Anal Biochem, 1976, 72:248-254.
pmid: 942051 |
| [19] |
Okibe N, Amada K, Hirano S, et al. Gene cloning and characterization of aldehyde dehydrogenase from a petroleum-degrading bacterium, strain HD-1[J]. J Biosci Bioeng, 1999, 88(1):7-11.
pmid: 16232565 |
| [20] |
Leal NA, Havemann GD, Bobik TA. PduP is a coenzyme-a-acylating propionaldehyde dehydrogenase associated with the polyhedral bodies involved in B12-dependent 1, 2-propanediol degradation by Salmonella enterica serovar Typhimurium LT2[J]. Arch Microbiol, 2003, 180(5):353-361.
doi: 10.1007/s00203-003-0601-0 URL |
| [21] | 赵振东, 李冬梅, 胡樨萼, 等. 抗松材线虫病马尾松种源化学成分与抗性机理研究(第Ⅱ报)- -马尾松种源抗性与中性萜类化合物组成差异关系研究[J]. 林产化学与工业, 2001, 21(1):56-60. |
| Zhao ZD, Li DM, Hu XE, et al. Study on chemical components and resistance mechanism to pine wood nematode of Masson pine provenance(ⅱ)-study on the components of neutral terpenoids and their differences among different resistant provenances of Pinus massoniana[J]. Chem Ind For Prod, 2001, 21(1):56-60. | |
| [22] | 喻方圆, 刘建兵, 胡晓健. 马尾松苗木中松醇的分布及其对干旱胁迫的响应[J]. 南京林业大学学报:自然科学版, 2009, 33(2):13-16. |
| Yu FY, Liu JB, Hu XJ. The distribution of pinitol in the Masson pine seedlings and its relation with drought stress[J]. J Nanjing For Univ Nat Sci Ed, 2009, 33(2):13-16. | |
| [23] | IKEDA T, ODA K. The occurrence of attractiveness for Monochamus alternatus hope(Coleoptera:Cerambycidae)in nematode-infected pine trees[J]. Nihon Ringakkai Shi/journal Jpn For Soc, 1980, 62:432-434. |
| [24] |
Zhao YH, Xu SY, Lu HB, et al. Effects of the plant volatile trans-2-hexenal on the dispersal ability, nutrient metabolism and enzymatic activities of Bursaphelenchus xylophilus[J]. Pestic Biochem Physiol, 2017, 143:147-153.
doi: 10.1016/j.pestbp.2017.08.004 URL |
| [25] |
Stiti N, Podgórska K, Bartels D. Aldehyde dehydrogenase enzyme ALDH3H1 from Arabidopsis thaliana:identification of amino acid residues critical for cofactor specificity[J]. Biochim Biophys Acta, 2014, 1844(3):681-693.
doi: 10.1016/j.bbapap.2014.01.008 pmid: 24463048 |
| [26] |
Corbier C, Della Seta F, Branlant G. A new chemical mechanism catalyzed by a mutated aldehyde dehydrogenase[J]. Biochemistry, 1992, 31(49):12532-12535.
pmid: 1463740 |
| [27] |
Schreiner L, Bauer P, Buettner A. Resolving the smell of wood - identification of odour-active compounds in Scots pine(Pinus sylvestris L.)[J]. Sci Rep, 2018, 8(1):8294.
doi: 10.1038/s41598-018-26626-8 pmid: 29844440 |
| [28] |
Kim JW, Im S, Jeong HR, et al. Neuroprotective effects of Korean red pine(Pinus densiflora)bark extract and its phenolics[J]. J Microbiol Biotechnol, 2018, 28(5):679-687.
doi: 10.4014/jmb.1801.01053 URL |
| [29] |
Li XM, Li YX, Wei DM, et al. Characterization of a broad-range aldehyde dehydrogenase involved in alkane degradation in Geobacillus thermodenitrificans NG80-2[J]. Microbiol Res, 2010, 165(8):706-712.
doi: 10.1016/j.micres.2010.01.006 URL |
| [1] | XIE Tian-peng, ZHANG Jia-ning, DONG Yong-jun, ZHANG Jian, JING Ming. Effect of Premature Bolting on the Rhizosphere Soil Microenvironment of Angelica sinensis [J]. Biotechnology Bulletin, 2023, 39(7): 206-218. |
| [2] | ZHAO Sai-sai, ZHANG Xiao-dan, JIA Xiao-yan, TAO Da-wei, LIU Ke-yu, NING Xi-bin. Investigation on the Complex Mutagenesis Selection of High-yield Nitrate Reductase Strain Staphylococcus simulans ZSJ6 and Its Enzymatic Properties [J]. Biotechnology Bulletin, 2023, 39(4): 103-113. |
| [3] | ZHANG Kai-ping, LIU Yan-li, TU Mian-liang, LI Ji-wei, WU Wen-biao. Optimization of Producing Cellulase by Aspergillus fumigatus A-16 and Its Enzymatic Properties [J]. Biotechnology Bulletin, 2022, 38(9): 215-225. |
| [4] | GAO Xiao-rong, DING Yao, LV Jun. Effects of Pseudomonas sp. PR3,a Pyrene-degrading Bacterium with Plant Growth-promoting Properties,on Rice Growth Under Pyrene Stress [J]. Biotechnology Bulletin, 2022, 38(9): 226-236. |
| [5] | QIN Xue-jing, WANG Yu-han, CAO Yi-bo, ZHANG Ling-yun. Prokaryotic Expression and Preparation of Polyclonal Antibody of PwHAP5 Gene in Picea wilsonii [J]. Biotechnology Bulletin, 2022, 38(8): 142-149. |
| [6] | HE Li-na, FENG Yuan, SHI Hui-min, YE Jian-ren. Screening and Identification of Endophytic Bacteria with Nematicidal Activity Against Bursaphelenchus xylophilus in Pinus massoniana [J]. Biotechnology Bulletin, 2022, 38(8): 159-166. |
| [7] | ZHAO Lin-yan, GUAN Hui-lin, WANG Ke-shu, LU Yan-lei, XIANG Ping, WEI Fu-gang, YANG Shao-zhou, XU Wu-mei. Effects of Soil Moisture on the Microbial Community Under Continuous Cropping of Panax notoginseng [J]. Biotechnology Bulletin, 2022, 38(7): 215-223. |
| [8] | YI Fang, LAI Peng-cheng, ZHENG Xi-ao, HU Shuai, GAO Yan-li. Research on the Preparation and Purification of Kod DNA Polymerase [J]. Biotechnology Bulletin, 2022, 38(5): 183-190. |
| [9] | WANG Xiao-qin, HUANG Yin-ping, WANG Wei-qian, WU Ping, QUAN Shu. Expression and Purification of the MLL3SET Protein with a Site-directed Mutation of an Unnatural Amino Acid [J]. Biotechnology Bulletin, 2022, 38(3): 194-202. |
| [10] | CHANG Qing, SHU Yue-rong, WANG Wen-tao, JIANG Hao, YAN Quan-de, QIAN Zheng, GAO Xue-chun, WU Jin-hong, ZHANG Yong. Heterologous Expression and Characterization of Endo-type Alginate Lyase from Yeosuana marina sp. JLT21 [J]. Biotechnology Bulletin, 2022, 38(2): 123-131. |
| [11] | ZHANG Feng-wen, ZHOU Li-ya, DONG Chao, SHI Yan-mao. Purification of Antioxidant Peptides from Natto Supernatant and Study on Its Activity [J]. Biotechnology Bulletin, 2022, 38(2): 158-165. |
| [12] | CHEN Duo, LIU Yong-zhe. Prokaryotic Expression,Purification and Crystallization of N-terminal Domain of Nucleocapsid Protein in SARS-CoV-2 [J]. Biotechnology Bulletin, 2022, 38(12): 149-155. |
| [13] | CAO Ru-fei, LI Ze-xuan, XU Huan, ZHANG Sha, ZHANG Min-min, DAI Feng, DUAN Xiao-lei. Expression,Purification,and Crystallization of Pif1 Helicase from Bacteroides fragilis [J]. Biotechnology Bulletin, 2021, 37(9): 180-190. |
| [14] | TIAN Jia-hui, FENG Jia-li, LU Jun-hua, MAO Lin-jing, HU Zhu-ran, WANG Ying, CHU Jie. Isolation,Purification and Characterization of Laccase LacT-1 from Cerrena unicolor [J]. Biotechnology Bulletin, 2021, 37(8): 186-194. |
| [15] | FANG Yuan, WU Xun, LIN Yu, WANG Hai-yan, WU Hui-ping, JU Yu-liang. Duplex-RPA Detection for Bursaphelenchus xylophilus and Bursaphelenchus mucronatus [J]. Biotechnology Bulletin, 2021, 37(7): 183-190. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||