








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (11): 137-149.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0686
Previous Articles Next Articles
HU Jin-chao( ), SHEN Wen-qi, XU Chao-ye, FAN Ya-qi, LU Hao-yu, JIANG Wen-jie, LI Shi-long, JIN Hong-chen, LUO Jian-mei(
), SHEN Wen-qi, XU Chao-ye, FAN Ya-qi, LU Hao-yu, JIANG Wen-jie, LI Shi-long, JIN Hong-chen, LUO Jian-mei( ), WANG Min
), WANG Min
Received:2023-07-15
Online:2023-11-26
Published:2023-12-20
Contact:
LUO Jian-mei
E-mail:15081112106@163.com;luojianmei@tust.edu.cn
HU Jin-chao, SHEN Wen-qi, XU Chao-ye, FAN Ya-qi, LU Hao-yu, JIANG Wen-jie, LI Shi-long, JIN Hong-chen, LUO Jian-mei, WANG Min. Research Advances in the Enhancement of Microbial Tolerance to Acid Stress[J]. Biotechnology Bulletin, 2023, 39(11): 137-149.
| 改造策略Modification strategy | 优点Advantages | 缺点Disadvantages |
|---|---|---|
| 适应性实验室进化 | 不需要了解菌株遗传信息、不依赖分子操作工具,可积累丰富的有益突变体 | 改造周期长、进化结果有一定随机性 |
| 预适应 | 耗时较短,可实现多种胁迫耐受性的交叉保护 | 适应结果随机性大,优良性状不稳定 |
| 基因组重排 | 不需要了解菌株遗传信息,不依赖分子操作工具,有利于多种优良性状的集合 | 对亲本菌株的质量要求较高,重排过程具有随机性,筛选过程费时费力 |
| 基因工程改造 | 靶向性高,便捷快速 | 高度依赖菌株的遗传背景和分子操作工具 |
| 全局转录机制工程 | 操作靶点少,能引发胞内的全局变化,适合复杂表型的改善 | 大量全局转录因子尚未被开发,改造靶向性差,筛选工作量大 |
| 系统生物学 | 改造通量大,可研究多个基因的协同作用,提升效果明显 | 多个基因的同时操作容易增加细胞的生理代谢负担 |
| 合成生物学 | 可根据环境变化对基因表达进行动态调控,减小细胞代谢负担 | 调控元件的挖掘不够广泛,复杂基因线路的导入可能与宿主代谢网络之间形成串扰 |
Table 1 Comparison of advantages and disadvantages of different modification strategies for enhancing acid resistance of microorganisms
| 改造策略Modification strategy | 优点Advantages | 缺点Disadvantages |
|---|---|---|
| 适应性实验室进化 | 不需要了解菌株遗传信息、不依赖分子操作工具,可积累丰富的有益突变体 | 改造周期长、进化结果有一定随机性 |
| 预适应 | 耗时较短,可实现多种胁迫耐受性的交叉保护 | 适应结果随机性大,优良性状不稳定 |
| 基因组重排 | 不需要了解菌株遗传信息,不依赖分子操作工具,有利于多种优良性状的集合 | 对亲本菌株的质量要求较高,重排过程具有随机性,筛选过程费时费力 |
| 基因工程改造 | 靶向性高,便捷快速 | 高度依赖菌株的遗传背景和分子操作工具 |
| 全局转录机制工程 | 操作靶点少,能引发胞内的全局变化,适合复杂表型的改善 | 大量全局转录因子尚未被开发,改造靶向性差,筛选工作量大 |
| 系统生物学 | 改造通量大,可研究多个基因的协同作用,提升效果明显 | 多个基因的同时操作容易增加细胞的生理代谢负担 |
| 合成生物学 | 可根据环境变化对基因表达进行动态调控,减小细胞代谢负担 | 调控元件的挖掘不够广泛,复杂基因线路的导入可能与宿主代谢网络之间形成串扰 |
| 菌株Strain | 靶点Targets | 提升效果Improving effect | 参考文献Reference |
|---|---|---|---|
| 大肠杆菌 Escherichia coli | 氢化酶辅助蛋白HypB和HypC | 乳酸产量提高2.1倍 | [ |
| 解脂耶氏酵母 Yarrowia lipolytica | 多效耐药蛋白 E25201g、膜组成蛋白 F16731g, B18854g、主要促进超家族渗透酶 F05984g | 阿魏酸耐受性从0.5 g/L提高到1.5 g/L | [ |
| 恶臭假单胞菌 Pseudomonas putida | 谷氨酸脱羧酶GadBC | pH 4.0下最终OD600提高30%以上 | [ |
| 大肠杆菌E. coli | 甲型流感基质-2(M2)蛋白 | 乙酸、庚酸、蓖麻油酸胁迫后CFU提高26.0%-65.0% | [ |
| 植物乳杆菌 Lactobacillus plantarum | 硫氧还原蛋白TrxA | pH 3.4下最终OD600提高40%左右 | [ |
| 植物乳杆菌L. plantarum | 转录因子CtsR | 酸-乙醇条件下最终OD600提高10%以上 | [ |
| 大肠杆菌E. coli | 转录因子H-NS | pH 2.5下存活性能提高100倍,pH 5.35乙酸和pH 5.35琥珀酸下生长性能提高30%和20%左右 | [ |
| 大肠杆菌E. coli | 过氧化氢酶KatE和超氧化物歧化酶SodB | 乙酰丙酸产量提高117.0% | [ |
| 酿酒酵母 Saccharomyces cerevisiae | 转录因子Haa1和磷酸核糖基焦磷酸合成酶PRS3 | 乙酸胁迫下葡萄糖消耗时间缩短70% | [ |
| 大肠杆菌E. coli | 整合宿主因子α亚基 IhfA、谷氨酸脱羧酶 GadA、DNA 修复蛋白 AidB 以及膜损伤蛋白 RyfA | 脂肪酸产量提高360% | [ |
| 大肠杆菌E. coli | 小分子RNA DsrA,分子伴侣Hfq | pH 2.5和pH4.5下存活率和最终OD600分别提高100倍和72% | [ |
| 巴氏醋杆菌 Acetobacter pasteurianus | 乙酰辅酶A合酶ACS1 | 乙酸产量提高153% | [ |
| 酿酒酵母S. cerevisiae | 乙酰辅酶A合酶ACS1 | 乙醇产量提高约10% | [ |
| 大肠杆菌E. coli | 周质伴侣HdeB、抗氧化酶SodB和KatE | pH 5.0下最终OD600提高57.0% | [ |
Table 2 Modification targets of cell tolerance to acids in this paper
| 菌株Strain | 靶点Targets | 提升效果Improving effect | 参考文献Reference |
|---|---|---|---|
| 大肠杆菌 Escherichia coli | 氢化酶辅助蛋白HypB和HypC | 乳酸产量提高2.1倍 | [ |
| 解脂耶氏酵母 Yarrowia lipolytica | 多效耐药蛋白 E25201g、膜组成蛋白 F16731g, B18854g、主要促进超家族渗透酶 F05984g | 阿魏酸耐受性从0.5 g/L提高到1.5 g/L | [ |
| 恶臭假单胞菌 Pseudomonas putida | 谷氨酸脱羧酶GadBC | pH 4.0下最终OD600提高30%以上 | [ |
| 大肠杆菌E. coli | 甲型流感基质-2(M2)蛋白 | 乙酸、庚酸、蓖麻油酸胁迫后CFU提高26.0%-65.0% | [ |
| 植物乳杆菌 Lactobacillus plantarum | 硫氧还原蛋白TrxA | pH 3.4下最终OD600提高40%左右 | [ |
| 植物乳杆菌L. plantarum | 转录因子CtsR | 酸-乙醇条件下最终OD600提高10%以上 | [ |
| 大肠杆菌E. coli | 转录因子H-NS | pH 2.5下存活性能提高100倍,pH 5.35乙酸和pH 5.35琥珀酸下生长性能提高30%和20%左右 | [ |
| 大肠杆菌E. coli | 过氧化氢酶KatE和超氧化物歧化酶SodB | 乙酰丙酸产量提高117.0% | [ |
| 酿酒酵母 Saccharomyces cerevisiae | 转录因子Haa1和磷酸核糖基焦磷酸合成酶PRS3 | 乙酸胁迫下葡萄糖消耗时间缩短70% | [ |
| 大肠杆菌E. coli | 整合宿主因子α亚基 IhfA、谷氨酸脱羧酶 GadA、DNA 修复蛋白 AidB 以及膜损伤蛋白 RyfA | 脂肪酸产量提高360% | [ |
| 大肠杆菌E. coli | 小分子RNA DsrA,分子伴侣Hfq | pH 2.5和pH4.5下存活率和最终OD600分别提高100倍和72% | [ |
| 巴氏醋杆菌 Acetobacter pasteurianus | 乙酰辅酶A合酶ACS1 | 乙酸产量提高153% | [ |
| 酿酒酵母S. cerevisiae | 乙酰辅酶A合酶ACS1 | 乙醇产量提高约10% | [ |
| 大肠杆菌E. coli | 周质伴侣HdeB、抗氧化酶SodB和KatE | pH 5.0下最终OD600提高57.0% | [ |
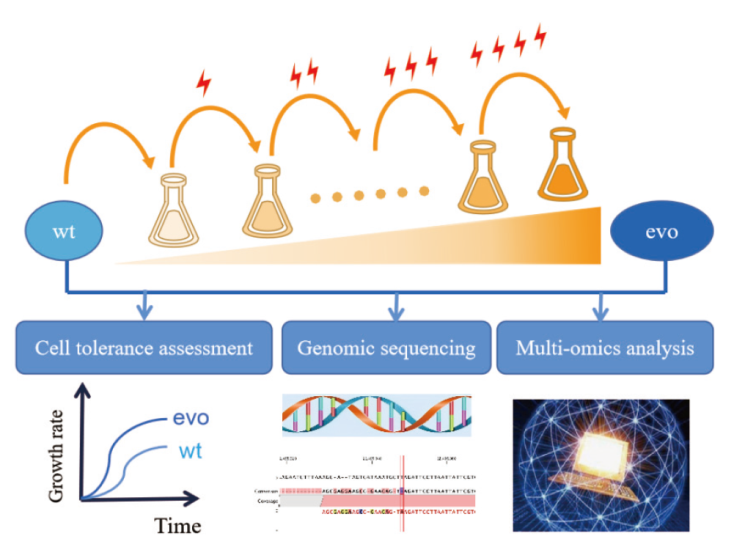
Fig. 1 Schematic representation of the enhancement on cell acid tolerance by the adaptive laboratory evolution wt: Wild-type strain; evo: evolutionary strain
| [1] | 朱亨政. 柠檬酸发酵[J]. 食品与发酵工业, 1994, 20(6): 69-74. |
| Zhu HZ. Citric acid fermentation[J]. Food Ferment Ind, 1994, 20(6): 69-74. | |
| [2] | 包启安. 醋酸菌及霉菌在食醋工业上的应用[J]. 食品科学, 1982, 3(6): 21-23. |
| Bao QA. Application of Acetobacter and mould in vinegar industry[J]. Food Sci, 1982, 3(6): 21-23. | |
| [3] |
Peleg Y, Rahamim E, Kessel M, et al. Malic acid accumulation by Aspergillus flavus[J]. Appl Microbiol Biotechnol, 1988, 28(1): 76-79.
doi: 10.1007/BF00250502 URL |
| [4] |
Guan NZ, Liu L. Microbial response to acid stress: mechanisms and applications[J]. Appl Microbiol Biotechnol, 2020, 104(1): 51-65.
doi: 10.1007/s00253-019-10226-1 pmid: 31773206 |
| [5] |
Yang JH, Peng Z, Zhu Q, et al. NiFe hydrogenase accessory proteins HypB-HypC accelerate proton conversion to enhance the acid resistance and d-lactic acid production of Escherichia coli[J]. ACS Synth Biol, 2022, 11(4): 1521-1530.
doi: 10.1021/acssynbio.1c00599 URL |
| [6] |
Zhang MH, Wu N, Fan YQ, et al. Proteomic profiling and stress response in Pediococcus acidilactici under acetic acid[J]. J Agric Food Chem, 2022, 70(39): 12708-12721.
doi: 10.1021/acs.jafc.2c04160 URL |
| [7] | 郝小明, 陈博, 安泰. 工业微生物酸胁迫的耐受机制及改造途径[J]. 生物工程学报, 2015, 31(8): 1151-1161. |
| Hao XM, Chen B, An T. Pathway modification of industrial microorganisms to improve acid-stress tolerance[J]. Chin J Biotechnol, 2015, 31(8): 1151-1161. | |
| [8] |
Radicella JP, Clark EA, Fox MS. Some mismatch repair activities in Escherichia coli[J]. Proc Natl Acad Sci USA, 1988, 85(24): 9674-9678.
doi: 10.1073/pnas.85.24.9674 pmid: 2974159 |
| [9] |
Ding J, Holzwarth G, Bradford CS, et al. PEP3 overexpression shortens lag phase but does not alter growth rate in Saccharomyces cerevisiae exposed to acetic acid stress[J]. Appl Microbiol Biotechnol, 2015, 99(20): 8667-8680.
doi: 10.1007/s00253-015-6708-9 pmid: 26051671 |
| [10] |
Lu PL, Ma D, Chen YL, et al. L-glutamine provides acid resistance for Escherichia coli through enzymatic release of ammonia[J]. Cell Res, 2013, 23(5): 635-644.
doi: 10.1038/cr.2013.13 |
| [11] | 张丽, 高健, 刘长青, 等. 耐受性工程调控微生物细胞工厂胁迫抗性[J]. 生物工程学报, 2022, 38(4): 1373-1389. |
| Zhang L, Gao J, Liu CQ, et al. Tolerance engineering regulates stress resistance of microbial cell factory[J]. Chin J Biotechnol, 2022, 38(4): 1373-1389. | |
| [12] |
Hu SM, You Y, Xia FF, et al. Genome shuffling improved acid-tolerance and succinic acid production of Actinobacillus succinogenes[J]. Food Sci Biotechnol, 2018, 28(3): 817-822.
doi: 10.1007/s10068-018-0505-z |
| [13] |
Sheehan VM, Sleator RD, Hill C, et al. Improving gastric transit, gastrointestinal persistence and therapeutic efficacy of the probiotic strain Bifidobacterium breve UCC2003[J]. Microbiol Read Engl, 2007, 153(Pt 10): 3563-3571.
doi: 10.1099/mic.0.2007/006510-0 URL |
| [14] |
Yan Z, Chen MX, Jia J, et al. Increasing acid tolerance of an engineered lactic acid bacterium Pediococcus acidilactici for L-lactic acid production[J]. Fermentation, 2022, 8(3): 96.
doi: 10.3390/fermentation8030096 URL |
| [15] |
Salas-Navarrete PC, de Oca Miranda AIM, Martínez A, et al. Evolutionary and reverse engineering to increase Saccharomyces cerevisiae tolerance to acetic acid, acidic pH, and high temperature[J]. Appl Microbiol Biotechnol, 2022, 106(1): 383-399.
doi: 10.1007/s00253-021-11730-z pmid: 34913993 |
| [16] |
Lubbers RJM, Liwanag AJ, Peng M, et al. Evolutionary adaptation of Aspergillus niger for increased ferulic acid tolerance[J]. J Appl Microbiol, 2020, 128(3): 735-746.
doi: 10.1111/jam.14505 pmid: 31674709 |
| [17] |
Wang ZD, Zhou LL, Lu MR, et al. Adaptive laboratory evolution of Yarrowia lipolytica improves ferulic acid tolerance[J]. Appl Microbiol Biotechnol, 2021, 105(4): 1745-1758.
doi: 10.1007/s00253-021-11130-3 |
| [18] |
Seong W, Han GH, Lim HS, et al. Adaptive laboratory evolution of Escherichia coli lacking cellular byproduct formation for enhanced acetate utilization through compensatory ATP consumption[J]. Metab Eng, 2020, 62: 249-259.
doi: 10.1016/j.ymben.2020.09.005 URL |
| [19] |
Almeida ELM, Ventorim RZ, Ferreira MAM, et al. New Papiliotrema laurentii UFV-1 strains with improved acetic acid tolerance selected by adaptive laboratory evolution[J]. Fungal Genet Biol, 2023, 164: 103765.
doi: 10.1016/j.fgb.2022.103765 URL |
| [20] |
Du B, Olson CA, Sastry AV, et al. Adaptive laboratory evolution of Escherichia coli under acid stress[J]. Microbiology, 2020, 166(2): 141-148.
doi: 10.1099/mic.0.000867 URL |
| [21] |
Ju SY, Kim JH, Lee PC. Long-term adaptive evolution of Leuconostoc mesenteroides for enhancement of lactic acid tolerance and production[J]. Biotechnol Biofuels, 2016, 9: 240.
doi: 10.1186/s13068-016-0662-3 URL |
| [22] | Overbeck TJ, Welker DL, Hughes JE, et al. Transient MutS-based hypermutation system for adaptive evolution of Lactobacillus casei to low pH[J]. Appl Environ Microbiol, 2017, 83(20): e01120-17. |
| [23] |
Nyabako BA, Fang H, Cui FJ, et al. Enhanced acid tolerance in Lactobacillus acidophilus by atmospheric and room temperature plasma(ARTP)coupled with adaptive laboratory evolution(ALE)[J]. Appl Biochem Biotechnol, 2020, 191(4): 1499-1514.
doi: 10.1007/s12010-020-03264-3 |
| [24] |
Zhao N, Zhang J, Qi YM, et al. New insights into thermo-acidophilic properties of Alicyclobacillus acidoterrestris after acid adaptation[J]. Food Microbiol, 2021, 94: 103657.
doi: 10.1016/j.fm.2020.103657 URL |
| [25] |
He GQ, Wu CD, Huang J, et al. Acid tolerance response of Tetragenococcus halophilus: a combined physiological and proteomic analysis[J]. Process Biochem, 2016, 51(2): 213-219.
doi: 10.1016/j.procbio.2015.11.035 URL |
| [26] |
Gu D, Wang KR, Lu TY, et al. Vibrio parahaemolyticus CadC regulates acid tolerance response to enhance bacterial motility and cytotoxicity[J]. J Fish Dis, 2021, 44(8): 1155-1168.
doi: 10.1111/jfd.v44.8 URL |
| [27] |
Yang H, Zhang L, Li JS, et al. Cell surface properties and transcriptomic analysis of cross protection provided between heat adaptation and acid stress in Tetragenococcus halophilus[J]. Food Res Int, 2021, 140: 110005.
doi: 10.1016/j.foodres.2020.110005 URL |
| [28] |
Huang M, Khan J, Kaur M, et al. CgSTE11 mediates cross tolerance to multiple environmental stressors in Candida glabrata[J]. Sci Rep, 2019, 9(1): 17036.
doi: 10.1038/s41598-019-53593-5 pmid: 31745168 |
| [29] |
Wu WQ, Pang B, Yang RR, et al. Improvement of the probiotic potential and yield of Lactobacillus rhamnosus cells using corn steep liquor[J]. LWT, 2020, 131: 109862.
doi: 10.1016/j.lwt.2020.109862 URL |
| [30] |
Wang WT, Wu B, Qin H, et al. Genome shuffling enhances stress tolerance of Zymomonas mobilis to two inhibitors[J]. Biotechnol Biofuels, 2019, 12: 288.
doi: 10.1186/s13068-019-1631-4 |
| [31] |
Zhou ZK, Liu YP, Zanaroli G, et al. Enhancing bioremediation potential of Pseudomonas putida by developing its acid stress tolerance with glutamate decarboxylase dependent system and global regulator of extreme radiation resistance[J]. Front Microbiol, 2019, 10: 2033.
doi: 10.3389/fmicb.2019.02033 URL |
| [32] |
Shin J, Jin YS, Park YC, et al. Enhancing acid tolerance of Escherichia coli via viroporin-mediated export of protons and its application for efficient whole-cell biotransformation[J]. Metab Eng, 2021, 67: 277-284.
doi: 10.1016/j.ymben.2021.07.007 URL |
| [33] |
Liu LX, Yu XY, Wu M, et al. Improved tolerance of Lactiplantiba-cillus plantarum in the presence of acid by the heterologous expression of trxA from Oenococcus oeni[J]. Fermentation, 2022, 8(9): 452.
doi: 10.3390/fermentation8090452 URL |
| [34] |
Zhao HY, Yuan L, Hu K, et al. Heterologous expression of ctsR from Oenococcus oeni enhances the acid-ethanol resistance of Lac-tobacillus plantarum[J]. FEMS Microbiol Lett, 2019, 366(15): fnz192.
doi: 10.1093/femsle/fnz192 URL |
| [35] |
Gao XX, Yang XF, Li JH, et al. Engineered global regulator H-NS improves the acid tolerance of E. coli[J]. Microb Cell Fact, 2018, 17(1): 118.
doi: 10.1186/s12934-018-0966-z |
| [36] |
Zhu CC, Chen JZ, Wang Y, et al. Enhancing 5-aminolevulinic acid tolerance and production by engineering the antioxidant defense system of Escherichia coli[J]. Biotechnol Bioeng, 2019, 116(8): 2018-2028.
doi: 10.1002/bit.v116.8 URL |
| [37] |
Cunha JT, Costa CE, Ferraz L, et al. HAA1 and PRS3 overexpression boosts yeast tolerance towards acetic acid improving xylose or glucose consumption: unravelling the underlying mechanisms[J]. Appl Microbiol Biotechnol, 2018, 102(10): 4589-4600.
doi: 10.1007/s00253-018-8955-z pmid: 29607452 |
| [38] |
Fang LX, Fan J, Luo SL, et al. Genome-scale target identification in Escherichia coli for high-titer production of free fatty acids[J]. Nat Commun, 2021, 12(1): 4976.
doi: 10.1038/s41467-021-25243-w |
| [39] | Lin ZL, Li JH, Yan XF, et al. Engineering of the small noncoding RNA(sRNA)DsrA together with the sRNA chaperone hfq enhances the acid tolerance of Escherichia coli[J]. Appl Environ Microbiol, 2021, 87(10): e02923-20. |
| [40] |
Zhu ZM, Yang PS, Yang JH, et al. Comparative transcriptome analysis reveals the contribution of membrane transporters to acid tolerance in Lactococcus lactis[J]. J Biotechnol, 2022, 357: 9-17.
doi: 10.1016/j.jbiotec.2022.08.006 URL |
| [41] |
Cao LY, Liu CG, Yang SH, et al. Regulation of biofilm formation in Zymomonas mobilis to enhance stress tolerance by heterologous expression of pfs and luxS[J]. Front Bioeng Biotechnol, 2023, 11: 1130405.
doi: 10.3389/fbioe.2023.1130405 URL |
| [42] |
Gao L, Wu XD, Li CY, et al. Exploitation of strong constitutive and stress-driven promoters from Acetobacter pasteurianus for improving acetic acid tolerance[J]. J Biotechnol, 2022, 350: 24-30.
doi: 10.1016/j.jbiotec.2022.03.013 URL |
| [43] |
Qin L, Dong SX, Yu J, et al. Stress-driven dynamic regulation of multiple tolerance genes improves robustness and productive capacity of Saccharomyces cerevisiae in industrial lignocellulose fermentation[J]. Metab Eng, 2020, 61: 160-170.
doi: 10.1016/j.ymben.2020.06.003 URL |
| [44] |
Yao XR, Liu P, Chen B, et al. Synthetic acid stress-tolerance modules improve growth robustness and lysine productivity of industrial Escherichia coli in fermentation at low pH[J]. Microb Cell Fact, 2022, 21(1): 68.
doi: 10.1186/s12934-022-01795-4 |
| [45] | 刘夏, 秦磊, 李春. 细胞工厂底盘抗逆属性的分子调控进展[J]. 生命科学, 2021, 33(12): 1452-1461. |
| Liu X, Qin L, Li C. Progress on molecular regulation of stress resistance properties for cell factory[J]. Chin Bull Life Sci, 2021, 33(12): 1452-1461. | |
| [46] | 李建, 孔婧, 李圣龙, 等. 适应性实验室进化技术在微生物育种中的应用进展[J]. 生物工程学报, 2021, 37(1): 130-141. |
| Li J, Kong J, Li SL, et al. Advances in adaptive laboratory evolutionary engineering to microbial breeding[J]. Chin J Biotechnol, 2021, 37(1): 130-141. | |
| [47] |
Sun HT, Lu ZN, Singh A, et al. Error-prone, stress-induced 3' flap-based Okazaki fragment maturation supports cell survival[J]. Science, 2021, 374(6572): 1252-1258.
doi: 10.1126/science.abj1013 pmid: 34855483 |
| [48] |
Kundu S, Peterson CL. Dominant role for signal transduction in the transcriptional memory of yeast GAL genes[J]. Mol Cell Biol, 2010, 30(10): 2330-2340.
doi: 10.1128/MCB.01675-09 URL |
| [49] |
Goudarzi A, Zhang D, Huang H, et al. Dynamic competing histone H4 K5 K8 acetylation and butyrylation are hallmarks of highly active gene promoters[J]. Mol Cell, 2016, 62(2): 169-180.
doi: S1097-2765(16)00224-0 pmid: 27105113 |
| [50] |
Tan YS, Zhang RK, Liu ZH, et al. Microbial adaptation to enhance stress tolerance[J]. Front Microbiol, 2022, 13: 888746.
doi: 10.3389/fmicb.2022.888746 URL |
| [51] | Liang RB, Swanson JMJ, Madsen JJ, et al. Acid activation mechanism of the influenza A M2 proton channel[J]. Proc Natl Acad Sci USA, 2016, 113(45): E6955-E6964. |
| [52] |
常瀚文, 郑鑫铃, 骆健美, 等. 抗逆元件及其在高效微生物细胞工厂构建中的应用进展[J]. 生物技术通报, 2020, 36(6): 13-34.
doi: 10.13560/j.cnki.biotech.bull.1985.2020-0259 |
| Chang HW, Zheng XL, Luo JM, et al. Tolerance elements and their application progress on the construction of highly-efficient microbial cell factory[J]. Biotechnol Bull, 2020, 36(6): 13-34. | |
| [53] |
周子康, 许平. 全局转录调控在细胞工厂构建中的应用与进展[J]. 化工进展, 2021, 40(3): 1248-1251.
doi: 10.16085/j.issn.1000-6613.2020-1907 |
|
Zhou ZK, Xu P. Application and progress of global transcription regulation in microbial cell factory construction[J]. Chem Ind Eng Prog, 2021, 40(3): 1248-1251.
doi: 10.16085/j.issn.1000-6613.2020-1907 |
|
| [54] |
Liu D, Hoynes-O'Connor A, Zhang FZ. Bridging the gap between systems biology and synthetic biology[J]. Front Microbiol, 2013, 4: 211.
doi: 10.3389/fmicb.2013.00211 pmid: 23898328 |
| [55] |
Battesti A, Majdalani N, Gottesman S. The RpoS-mediated general stress response in Escherichia coli[J]. Annu Rev Microbiol, 2011, 65: 189-213.
doi: 10.1146/annurev-micro-090110-102946 pmid: 21639793 |
| [56] |
Gottesman S. Trouble is coming: Signaling pathways that regulate general stress responses in bacteria[J]. J Biol Chem, 2019, 294(31): 11685-11700.
doi: 10.1074/jbc.REV119.005593 pmid: 31197038 |
| [57] |
Wang WW, Wang LJ, Wu JH, et al. Hfq-bridged ternary complex is important for translation activation of rpoS by DsrA[J]. Nucleic Acids Res, 2013, 41(11): 5938-5948.
doi: 10.1093/nar/gkt276 pmid: 23605038 |
| [58] | 严伟, 信丰学, 董维亮, 等. 合成生物学及其研究进展[J]. 生物学杂志, 2020, 37(5): 1-9. |
| Yan W, Xin FX, Dong WL, et al. Synthetic biology and research progress[J]. J Biol, 2020, 37(5): 1-9. | |
| [59] |
Xiong L, Zeng Y, Tang RQ, et al. Condition-specific promoter activities in Saccharomyces cerevisiae[J]. Microb Cell Fact, 2018, 17(1): 58.
doi: 10.1186/s12934-018-0899-6 |
| [60] | Yin X, Shin HD, Li JH, et al. P gas, a low-pH-induced promoter, as a tool for dynamic control of gene expression for metabolic engineering of Aspergillus niger[J]. Appl Environ Microbiol, 2017, 83(6): e03222-16. |
| [61] |
Khalil AS, Collins JJ. Synthetic biology: applications come of age[J]. Nat Rev Genet, 2010, 11(5): 367-379.
doi: 10.1038/nrg2775 pmid: 20395970 |
| [62] |
许可, 王靖楠, 李春. 智能抗逆微生物细胞工厂与绿色生物制造[J]. 合成生物学, 2020, 1(4): 427-439.
doi: 10.12211/2096-8280.2020-045 |
| Xu K, Wang JN, Li C. Intelligent microbial cell factory with tolerance for green biological manufacturing[J]. Synth Biol J, 2020, 1(4): 427-439. | |
| [63] |
Mira NP, Becker JD, Sá-Correia I. Genomic expression program involving the Haa1p-regulon in Saccharomyces cerevisiae response to acetic acid[J]. OMICS, 2010, 14(5): 587-601.
doi: 10.1089/omi.2010.0048 URL |
| [1] | ZHANG Kun, YAN Chang, TIAN Xin-peng. Research Progress in Microbial Single Cell Separation Methods [J]. Biotechnology Bulletin, 2023, 39(9): 1-11. |
| [2] | ZHAO Lin-yan, XU Wu-mei, WANG Hao-ji, WANG Kun-yan, WEI Fu-gang, YANG Shao-zhou, GUAN Hui-lin. Effects of Applying Biochar on the Rhizosphere Fungal Community and Survival Rate of Panax notoginseng Under Continuous Cropping [J]. Biotechnology Bulletin, 2023, 39(7): 219-227. |
| [3] | WAN Qi-wu, BAO Xu-dong, DING Ke, MOU Hua-ming, LUO Yang. Research Progress in Microfluidic Technology in the Detection of Pathogenic Microorganisms [J]. Biotechnology Bulletin, 2023, 39(10): 107-114. |
| [4] | YANG Lu, XIN Jian-pan, TIAN Ru-nan. Research Progress in the Mitigative Effects of Rhizosphere Microorganisms on Heavy Metal Stress in Plants and Their Mechanisms [J]. Biotechnology Bulletin, 2022, 38(3): 213-225. |
| [5] | WU Qi-man, ZHANG Jin-mei, LI Yue-ying, ZHANG Ying. Recent Advances on the Mechanism of Beneficial Microbial Fertilizers in Crops [J]. Biotechnology Bulletin, 2021, 37(5): 221-230. |
| [6] | HUANG Hai-min, LAN Xiu-wan, WU Yao-sheng. Research Progress on Intestinal Microbe and Sex Hormone Related Diseases [J]. Biotechnology Bulletin, 2020, 36(2): 77-82. |
| [7] | LI Jian-tao, LIU Xian-hua, HE Yao-dong, WANG Guang-yi. Mutagenesis Breeding in DHA Production by Oleaginous Microorganisms [J]. Biotechnology Bulletin, 2020, 36(1): 110-115. |
| [8] | ZHOU Heng, JIANG Yun, XU Ye-xiang, QIAN Sheng-hui, MIAO Li. Research on Quorum Sensing Inhibitory Activity and Culture Condition of a Marine Streptomyces parvulus [J]. Biotechnology Bulletin, 2019, 35(10): 137-143. |
| [9] | XU Shan ,LI Ren-qiang ,ZHENG Zhen-hua ,ZHANG Yun ,SUN Ai-jun ,HU Yun-feng. Properties of Extracellular Protease of Microbe DH-2 from Mangrove and Optimization of Enzyme Producing Conditions [J]. Biotechnology Bulletin, 2018, 34(6): 120-127. |
| [10] | WANG Da-zhou ,GUO Tian-xiao ,ZHENG Shi, SHANG Ying, XU Wen-tao. Research Progress on the Isothermal Nucleic Acid Amplification Techniques in Rapid Detection of Microorganisms [J]. Biotechnology Bulletin, 2017, 33(7): 49-61. |
| [11] | LIANG Jin-gang, ZHANG Xiu-jie. Strategies for Evaluating the Effects of Transgenic Crops on Soil Microbial Diversity [J]. Biotechnology Bulletin, 2017, 33(10): 111-116. |
| [12] | ZHAO Wei-jun, HUANG Lei, XU Zhi-nan. Research Progress on the Synthesis of S-adenosyl-L-methionine in Microorganism [J]. Biotechnology Bulletin, 2017, 33(1): 99-105. |
| [13] | Bao Hanyan, Gu Pengjuan, Zhang Yi, Mu Jun, Feng Yan, Zhao Chenyan. Screening the Extracts of Marine Fungi for the Activity of Damaging the DNA of Escherichia coli and Researches on Active Strains [J]. Biotechnology Bulletin, 2015, 31(8): 132-139. |
| [14] | Liao Qing, Jiang Zhepu, Xingying, Wei Guangpo, Huang Dongliang, Li Yangrui. Isolation and Identification of Cellulose-Decomposing Microorganisms in Deep-litter System [J]. Biotechnology Bulletin, 2014, 0(3): 106-110. |
| [15] | Feng Xuezhen, Wu Shanguang, Lu Yuan. Preliminary Application of PCR-DGGE to Analyzing Microbial Diversity of Ulva lactuca L. and Dictyota dichotoma [J]. Biotechnology Bulletin, 2014, 0(12): 73-77. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||