








Biotechnology Bulletin ›› 2023, Vol. 39 ›› Issue (8): 220-233.doi: 10.13560/j.cnki.biotech.bull.1985.2023-0115
Previous Articles Next Articles
LIU Bao-cai1,2( ), CHEN Jing-ying1,2(
), CHEN Jing-ying1,2( ), ZHANG Wu-jun1,2, HUANG Ying-zhen1,2, ZHAO Yun-qing1,2, LIU Jian-chao3, WEI Zhi-cheng4
), ZHANG Wu-jun1,2, HUANG Ying-zhen1,2, ZHAO Yun-qing1,2, LIU Jian-chao3, WEI Zhi-cheng4
Received:2023-02-13
Online:2023-08-26
Published:2023-09-05
Contact:
CHEN Jing-ying
E-mail:626813844@qq.com;cjy6601@163.com
LIU Bao-cai, CHEN Jing-ying, ZHANG Wu-jun, HUANG Ying-zhen, ZHAO Yun-qing, LIU Jian-chao, WEI Zhi-cheng. Characteristics Analysis of Seed Microrhizome Gene Expression of Polygonatum cyrtonema[J]. Biotechnology Bulletin, 2023, 39(8): 220-233.

Fig. 1 Germination process of P. cyrtonema seeds A: Radicle breaks through seed coat; B: radicle elongation; C: hypocotyl enlargement; D: microrhizomes; E: green microrhizomes; F: seedlings. A, D and E in germination stage were used as experimental materials in this experiment
| 基因 Gene | 引物 Primer | 序列 Sequence(5'-3') |
|---|---|---|
| AUX1 | Cluster-68615.91119-F | CCTCTCCTTCTTGGTCCTGTA |
| Cluster-68615.91119-R | GGTGGCTCAACTTATGATGCT | |
| AUX2 | Cluster-68615.87861-F | TGCTCATCCATCAGTTCATAACC |
| Cluster-68615.87861-R | GAATACGATTGCGAGGAACCATA | |
| CH3 | Cluster-58608.0-F | AAGAAGACCTCCAGAAGAGCATA |
| Cluster-58608.0-R | GACAATCTCCCAGAACAACACAT | |
| TF | Cluster-68615.45537-F | CTTCCTGCTGCTTTCTCTTAGTG |
| Cluster-68615.45537-R | TGCGGCTGCTCAGATTATTG | |
| SnPK2 | Cluster-68615.69985-F | ATGCTGATGACTCTGATTCTGATG |
| Cluster-68615.69985-R | GCCTACAACCGATACTTCTGATAC | |
| MYC2 | Cluster-68615.81195-F | AGATCAGCTCAGCTTCCATCA |
| Cluster-68615.81195-R | CATTCCAGTTCCTTCGCCATT | |
| β-glucosidase | Cluster-68615.43996-F | ACAAGGGTTAATGGGACTTCTCT |
| Cluster-68615.43996-R | CTGAAGAATCGCCTCCAAGTG | |
| Actin | Actin_Cluster-F | CACCGATTGACACAAGGAGAG |
| Actin_Cluster-R | AGGATGGCTTACTACATTGACTTC |
Table 1 Gene-specific primers used in RT-qPCR
| 基因 Gene | 引物 Primer | 序列 Sequence(5'-3') |
|---|---|---|
| AUX1 | Cluster-68615.91119-F | CCTCTCCTTCTTGGTCCTGTA |
| Cluster-68615.91119-R | GGTGGCTCAACTTATGATGCT | |
| AUX2 | Cluster-68615.87861-F | TGCTCATCCATCAGTTCATAACC |
| Cluster-68615.87861-R | GAATACGATTGCGAGGAACCATA | |
| CH3 | Cluster-58608.0-F | AAGAAGACCTCCAGAAGAGCATA |
| Cluster-58608.0-R | GACAATCTCCCAGAACAACACAT | |
| TF | Cluster-68615.45537-F | CTTCCTGCTGCTTTCTCTTAGTG |
| Cluster-68615.45537-R | TGCGGCTGCTCAGATTATTG | |
| SnPK2 | Cluster-68615.69985-F | ATGCTGATGACTCTGATTCTGATG |
| Cluster-68615.69985-R | GCCTACAACCGATACTTCTGATAC | |
| MYC2 | Cluster-68615.81195-F | AGATCAGCTCAGCTTCCATCA |
| Cluster-68615.81195-R | CATTCCAGTTCCTTCGCCATT | |
| β-glucosidase | Cluster-68615.43996-F | ACAAGGGTTAATGGGACTTCTCT |
| Cluster-68615.43996-R | CTGAAGAATCGCCTCCAAGTG | |
| Actin | Actin_Cluster-F | CACCGATTGACACAAGGAGAG |
| Actin_Cluster-R | AGGATGGCTTACTACATTGACTTC |
| 样品 Sample | 总读长 Total reads/bp | 总匹配数 Total mapped ones/bp |
|---|---|---|
| A1 | 40 179 174 | 29 824 074(74.23%) |
| A2 | 45 826 078 | 32 824 940(71.63%) |
| A3 | 44 560 782 | 32 498 548(72.93%) |
| D1 | 45 873 682 | 34 073 516(74.28%) |
| D2 | 41 999 498 | 31 537 406(75.09%) |
| D3 | 45 760 904 | 33 679 092(73.60%) |
| E1 | 41 290 430 | 30 888 404(74.81%) |
| E2 | 44 373 676 | 33 739 212(76.03%) |
| E3 | 44 063 488 | 33 299 498(75.57%) |
Table 2 Mapped results of sample reads and assembly transcripts
| 样品 Sample | 总读长 Total reads/bp | 总匹配数 Total mapped ones/bp |
|---|---|---|
| A1 | 40 179 174 | 29 824 074(74.23%) |
| A2 | 45 826 078 | 32 824 940(71.63%) |
| A3 | 44 560 782 | 32 498 548(72.93%) |
| D1 | 45 873 682 | 34 073 516(74.28%) |
| D2 | 41 999 498 | 31 537 406(75.09%) |
| D3 | 45 760 904 | 33 679 092(73.60%) |
| E1 | 41 290 430 | 30 888 404(74.81%) |
| E2 | 44 373 676 | 33 739 212(76.03%) |
| E3 | 44 063 488 | 33 299 498(75.57%) |
| 样品 Sample | FPKM 区间FPKM Interval | ||||||
|---|---|---|---|---|---|---|---|
| 0≤FPKM≤0.1 | 0.1<FPKM≤0.3 | 0.3<FPKM≤3.57 | 3.57<FPKM≤15 | 15<FPKM≤60 | FPKM>60 | ||
| A1 | 45 729(25.64%) | 8 928(5.01%) | 84 666(47.48%) | 26 418(14.82%) | 9 731(5.46%) | 2 847(1.60%) | |
| A2 | 90 461(50.73%) | 4 155(2.33%) | 46 585(26.12%) | 25 282(14.18%) | 8 776(4.92%) | 3 060(1.72%) | |
| A3 | 92 713(51.99%) | 2 373(1.33%) | 42 307(23.73%) | 28 240(15.84%) | 9 676(5.43%) | 3 010(1.69%) | |
| D1 | 68 567(38.45%) | 14 059(7.88%) | 64 246(36.03%) | 19 173(10.75%) | 9 466(5.31%) | 2 808(1.57%) | |
| D2 | 68 767(38.56%) | 13 366(7.50%) | 64 688(36.28%) | 19 321(10.84%) | 9 411(5.28%) | 2 766(1.55%) | |
| D3 | 69 033(38.71%) | 13 351(7.49%) | 64 127(35.96%) | 19 404(10.88%) | 9 570(5.37%) | 2 834(1.59%) | |
| E1 | 63 577(35.65%) | 11 564(6.49%) | 70 391(39.47%) | 20 459(11.47%) | 9 569(5.37%) | 2 759(1.55%) | |
| E2 | 61 068(34.25%) | 13 276(7.45%) | 72 028(40.39%) | 19 855(11.13%) | 9 390(5.27%) | 2 702(1.52%) | |
| E3 | 59 900(33.59%) | 13 120(7.36%) | 73 014(40.95%) | 20 113(11.28%) | 9 453(5.30%) | 2 719(1.52%) | |
Table 3 FPKM interval statistics of sample expressed levels
| 样品 Sample | FPKM 区间FPKM Interval | ||||||
|---|---|---|---|---|---|---|---|
| 0≤FPKM≤0.1 | 0.1<FPKM≤0.3 | 0.3<FPKM≤3.57 | 3.57<FPKM≤15 | 15<FPKM≤60 | FPKM>60 | ||
| A1 | 45 729(25.64%) | 8 928(5.01%) | 84 666(47.48%) | 26 418(14.82%) | 9 731(5.46%) | 2 847(1.60%) | |
| A2 | 90 461(50.73%) | 4 155(2.33%) | 46 585(26.12%) | 25 282(14.18%) | 8 776(4.92%) | 3 060(1.72%) | |
| A3 | 92 713(51.99%) | 2 373(1.33%) | 42 307(23.73%) | 28 240(15.84%) | 9 676(5.43%) | 3 010(1.69%) | |
| D1 | 68 567(38.45%) | 14 059(7.88%) | 64 246(36.03%) | 19 173(10.75%) | 9 466(5.31%) | 2 808(1.57%) | |
| D2 | 68 767(38.56%) | 13 366(7.50%) | 64 688(36.28%) | 19 321(10.84%) | 9 411(5.28%) | 2 766(1.55%) | |
| D3 | 69 033(38.71%) | 13 351(7.49%) | 64 127(35.96%) | 19 404(10.88%) | 9 570(5.37%) | 2 834(1.59%) | |
| E1 | 63 577(35.65%) | 11 564(6.49%) | 70 391(39.47%) | 20 459(11.47%) | 9 569(5.37%) | 2 759(1.55%) | |
| E2 | 61 068(34.25%) | 13 276(7.45%) | 72 028(40.39%) | 19 855(11.13%) | 9 390(5.27%) | 2 702(1.52%) | |
| E3 | 59 900(33.59%) | 13 120(7.36%) | 73 014(40.95%) | 20 113(11.28%) | 9 453(5.30%) | 2 719(1.52%) | |
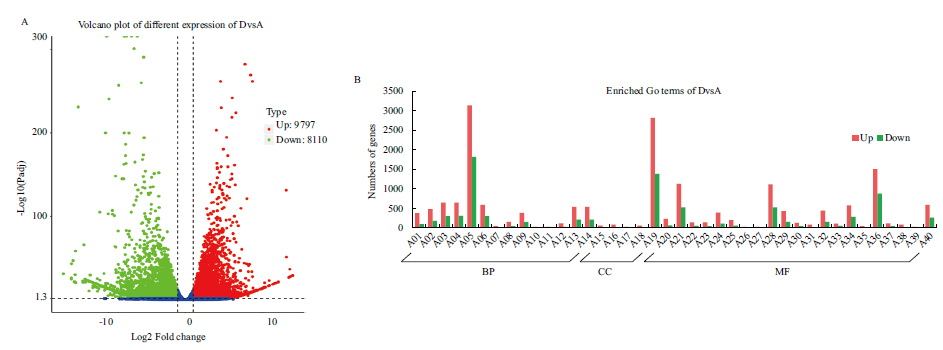
Fig. 2 DvsA different expressed genes A01: Protein phosphorylation. A02: Phosphorylation. A03: Phosphate-containing compound metabolic process. A04: Phosphorus metabolic process. A05: Metabolic process. A06: Oxidation-reduction process. A07: Microtubule-based movement. A08: Cellular carbohydrate metabolic process. A09: Carbohydrate metabolic process. A10: Negative regulation of translation. A11: Negative regulation of cellular amide metabolic process. A12: Cellular polysaccharide metabolic process. A13: Cellular protein modification process. A14: Protein modification process. A15: Microtubule. A16: Tubulin complex. A17: Apoplast. A18: Cell wall. A19: Catalytic activity. A20: Hydrolase activity, acting on glycosyl bonds. A21:Transferase activity. A22: Tetrapyrrole binding, A23: Heme binding. A24: Protein kinase activity. A25: Hydrolase activity, hydrolyzing O-glycosyl compounds. A26: RNA glycosylase activity. A27: rRNA N-glycosylase activity. A28: Hydrolase activity. A29: Phosphotransferase activity, alcohol group as acceptor. A30: Oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen. A31: Microtubule binding. A32: Kinase activity. A33: Transferase activity, transferring acyl groups other than amino-acyl groups. A34: Oxidoreductase activity. A35: Microtubule motor activity. A36: Ion binding. A37: Iron ion binding. A38: Tubulin binding. A39: Xyloglucan: xyloglucosyl transferase activity. A40: Transferase activity, transferring phosphorus-containing groups
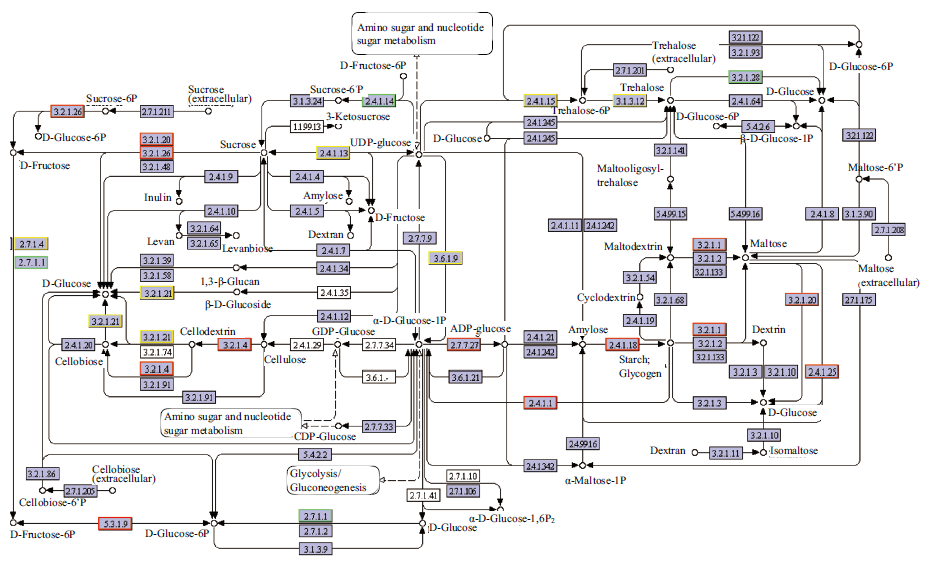
Fig. 5 DvsA differentially expressed genes involved in starch and sucrose metabolism The red border of the node contains up-regulated differential genes; the green border of the node contains the down-regulated differential genes; and the yellow border of the node contains the up-regulated and down-regulated differential genes. The same below

Fig. 7 EvsD differently expressed genes B01: Ribosome biogenesis. B02: Translation. B03: Ribonucleoprotein complex biogenesis. B04: Peptide biosynthetic process. B05: Peptide metabolic process. B06: Cellular amide metabolic process. B07: Amide biosynthetic process. B08: Oxidation-reduction process. B09: Organonitrogen compound biosynthetic process. B10: Organonitrogen compound metabolic process. B11: Cellular component biogenesis. B12: Protein metabolic process. B13: Metabolic process. B14: Cellular protein metabolic process. B15: Biosynthetic process. B16: Organic substance biosynthetic process. B17: Cellular macromolecule biosynthetic process. B18: Cellular component organization or biogenesis. B19: Macromolecule biosynthetic process. B20: Cellular nitrogen compound biosynthetic process. B21: Cellular biosynthetic process. B22: Single-organism metabolic process. B23: Obsolete peroxidase reaction. B24: Ribosome. B25: Ribonucleoprotein complex. B26: Non-membrane-bounded organelle. B27: Intracellular non-membrane-bounded organelle. B28: Cytoplasmic part. B29: Cytoplasm. B30: Macromolecular complex. B31: Structural constituent of ribosome. B32: Oxidoreductase activity. B33: Structural molecule activity. B34: Catalytic activity. B35: Antioxidant activity. B36: Oxidoreductase activity, acting on peroxide as acceptor. B37: Pyrophosphatase activity. B38: Peroxidase activity. B39: Nucleoside-triphosphatase activity. B40: GTPase activity.

Fig. 11 Verification of seven selected DEGs by RT-qPCR Comparison of RNA-seq data(blue line chart)with RT-qPCR data(orange bar graph). Different lowercase letters indicate significant differences in the expression levels of the same gene in different morphology of micro-rhizome development at 0.05 level
| [34] | Shen FM, Xie P, Li CT, et al. Polysaccharides from Polygonatum cyrtonema Hua reduce depression-like behavior in mice by inhibiting oxidative stress-calpain-1-NLRP3 signaling axis[J]. Oxid Med Cell Longev, 2022, 2022: 2566917. |
| [1] |
Nabuuma D, Reimers C, Hoang KT, et al. Impact of seed system interventions on food and nutrition security in low- and middle-income countries: a scoping review[J]. Glob Food Secur, 2022, 33: 100638.
doi: 10.1016/j.gfs.2022.100638 URL |
| [2] | 徐恒恒, 黎妮, 刘树君, 等. 种子萌发及其调控的研究进展[J]. 作物学报, 2014, 40(7): 1141-1156. |
|
Xu HH, Li N, Liu SJ, et al. Research progress in seed germination and its control[J]. Acta Agron Sin, 2014, 40(7): 1141-1156.
doi: 10.3724/SP.J.1006.2014.01141 URL |
|
| [3] |
Han C, Yang PF. Studies on the molecular mechanisms of seed germination[J]. Proteomics, 2015, 15(10): 1671-1679.
doi: 10.1002/pmic.201400375 pmid: 25597791 |
| [4] | 王剑龙, 常晖, 周仔莉, 等. 黄精种子萌发过程发育解剖学研究[J]. 西北植物学报, 2013, 33(8): 1584-1588 |
| Wang JL, Chang H, Zhou ZL, et al. Developmental anatomy of Polygonatum sibiricum red.during the seed germination process[J]. Acta Bot Boreali Occidentalia Sin, 2013, 33(8): 1584-1588 | |
| [5] | 陈怡, 杨赋祺, 陈松树, 等. 多花黄精种子萌发过程的内源激素含量变化研究[J]. 中药材, 2020, 43(3): 523-527. |
| Chen Y, Yang FQ, Chen SS, et al. Study on the changes of endogenous hormones contents in Polygonatum cyrtonema seed germination[J]. J Central Univ Finance Econ, 2020, 43(3): 523-527. | |
| [6] | Tang ZB, Zhao J, Yang B, et al. Dynamic RNA-seq study reveals the potential regulators of seed germination in Paris polyphylla var. yunnanensis[J]. Plants(Basel), 2022, 11(18): 2400. |
| [7] | 杨瑞霜, 吕昕芮, 刘祖懿, 等. 七叶一枝花种子萌发过程的形态及皂苷类物质含量变化[J/OL]. 分子植物育种, 2022. https://kns.cnki.net/kcms/detail/46.1068.S.20220307.1054.008.html. |
| Yang RS, Lyu XR, Liu ZY, et al. The Changes of morphology and saponins content during paris polyphylla var. chinensis seeds germination[J/OL]. Mol Plant Breed, 2022. https://kns.cnki.net/kcms/detail/46.1068.S.20220307.1054.008.html. | |
| [8] | 陈怡, 柳雪晨, 陈松树, 等. 多花黄精种子萌发过程的形态和解剖研究[J]. 种子, 2020, 39(2): 5-10 |
| Chen Y, Liu XC, Chen SS, et al. Morphological and anatomical studies during seed germination of Polygonatum cyrtonema Hua[J]. Seed, 2020, 39(2): 5-10 | |
| [9] |
Ling LZ, Zhang SD. Comparative proteomic analysis between mature and germinating seeds in Paris polyphylla var. yunnanensis[J]. PeerJ, 2022, 10(2): e13304.
doi: 10.7717/peerj.13304 URL |
| [10] | Chen XQ, Liang SY, Xu JM. Polygonatum cyrtonema[M]// Flora of China. Louis: Missouri Botanical Garden Press, 2000, 24: 229 |
| [11] | 刘保财, 黄颖桢, 赵云青, 等. 多花黄精种苗繁育技术[J]. 福建农业科技, 2017(9): 36-38. |
| Liu BC, Huang YZ, Zhao YQ, et al. Seedling multiplication technology of Polygonatum cyrtonema Hua[J]. Fujian Agric Sci Technol, 2017(9): 36-38. | |
| [12] | 斯金平, 朱玉贤. 黄精——一种潜力巨大且不占农田的新兴优质杂粮[J]. 中国科学: 生命科学, 2021, 51(11): 1477-1484. |
|
Si JP, Zhu YX. Polygonatum sibiricum—a kind of emerging high-quality coarse cereals with great potential and not occupying farmland[J]. Sci Sin Vitae, 2021, 51(11): 1477-1484.
doi: 10.1360/SSV-2020-0413 URL |
|
| [13] |
Li B, Dewey C. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome[J]. BMC Bioinform, 2011, 12(1): 93-99.
doi: 10.1186/1471-2105-12-93 |
| [14] |
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2[J]. Genome Biol, 2014, 15(12): 550.
doi: 10.1186/s13059-014-0550-8 URL |
| [15] |
Young M, Wakefield M, Smyth G, et al. Gene ontology analysis for RNA-seq: accounting for selection bias[J]. Genome Biol, 2010, 11(2): R14.
doi: 10.1186/gb-2010-11-2-r14 URL |
| [16] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T))[J]. Methods, 2001, 25(4): 402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [17] | 王晶晶, 时金殿, 李晨萱, 等. 多花黄精和滇黄精种子的营养成分及抗氧化物酶活性[J]. 贵州农业科学, 2022, 50(1): 90-95. |
| Wang JJ, Shi JD, Li CX, et al. Nutritional components and antioxidant enzyme activity of Polygonatum cyrtonema and Polygonatum kingianum seeds[J]. Guizhou Agric Sci, 2022, 50(1): 90-95. | |
| [18] |
Xue T, Zhao MH, Chen J, et al. Revealing the mechanisms of the bioactive ingredients accumulation in Polygonatum cyrtonema by multiomics analyses[J]. Front Plant Sci, 2022, 13: 1055721.
doi: 10.3389/fpls.2022.1055721 URL |
| [19] |
Qi JJ, Wei JH, Liao DQ, et al. Untargeted metabolomics analysis revealed the major metabolites in the seeds of four Polygonatum species[J]. Molecules, 2022, 27(4): 1445.
doi: 10.3390/molecules27041445 URL |
| [20] | 张武君, 赵云青, 刘保财, 等. 多花黄精种子层积过程生理变化研究[J]. 福建农业学报, 2022, 37(8): 995-1007. |
| Zhang WJ, Zhao YQ, Liu BC, et al. Physiological changes of Polygonatum cyrtonema Hua seeds during stratification[J]. Fujian J Agric Sci, 2022, 37(8): 995-1007. | |
| [21] |
Huang SH, Zheng CY, Zhao Y, et al. RNA interference knockdown of the brassinosteroid receptor BRI1 in potato(Solanum tuberosum L.) reveals novel functions for brassinosteroid signaling in controlling tuberization[J]. Sci Hortic, 2021, 290: 110516.
doi: 10.1016/j.scienta.2021.110516 URL |
| [22] |
罗丽娜, 向增旭. 基于转录组测序分析的黄精种子休眠解除相关差异基因研究[J]. 中国农学通报, 2021, 37(11): 1-8.
doi: 10.11924/j.issn.1000-6850.casb2020-0209 |
| Luo LN, Xiang ZX. The relative difference genes in the process of dormancy release of Polygonatum sibiricum Red.Based on the transcriptome sequencing analysis[J]. Chin Agric Sci Bull, 2021, 37(11): 1-8. | |
| [23] |
Liao DQ, An RP, Wei JH, et al. Transcriptome profiles revealed molecular mechanisms of alternating temperatures in breaking the epicotyl morphophysiological dormancy of Polygonatum sibiricum seeds[J]. BMC Plant Biol, 2021, 21(1): 370.
doi: 10.1186/s12870-021-03147-7 |
| [24] | 周新华, 桂尚上, 肖智勇, 等. 温度和光照对多花黄精种子萌发的影响[J]. 南方林业科学, 2016, 44(6): 1-4, 26 |
| Zhou XH, Gui SS, Xiao ZY, et al. Influence of temperature and light on the seed germination of Polygonatum cyrtonema[J]. South China For Sci, 2016, 44(6): 1-4, 26 | |
| [25] | 刘保财, 黄颖桢, 赵云青, 等. 不同处理对多花黄精种子的影响[J]. 福建农业学报, 2015, 30(5): 469-472. |
| Liu BC, Huang YZ, Zhao YQ, et al. Effect of varied treatments on germination of Polygonatum cyrtonema Hua seeds[J]. Fujian J Agric Sci, 2015, 30(5): 469-472. | |
| [26] |
Zhang WW, Xia L, Peng FL, et al. Transcriptomics and metabolomics changes triggered by exogenous 6-benzylaminopurine in relieving epicotyl dormancy of Polygonatum cyrtonema Hua seeds[J]. Front Plant Sci, 2022, 13: 961899.
doi: 10.3389/fpls.2022.961899 URL |
| [27] |
杜佳慧, 徐伟芳, 杨晓冬, 等. 多花黄精产吲哚乙酸内生菌的分离筛选及其对黄精种子萌发的影响[J]. 生物技术通报, 2022, 38(12): 223-232.
doi: 10.13560/j.cnki.biotech.bull.1985.2022-0078 |
| Du JH, Xu WF, Yang XD, et al. Isolation and screening of endophytes producing indole acetic acid from Polygonatum cyrtonema Hua. and its effect on seed germination of Polygonatum[J]. Biotechnol Bull, 2022, 38(12): 223-232. | |
| [28] |
骆绪美, 许早时, 韩文妍, 等. 多花黄精实生育苗及幼苗生物质积累规律分析[J]. 农学学报, 2021, 11(11): 76-80
doi: 10.11923/j.issn.2095-4050.cjas2020-0007 |
| Luo XM, Xu ZS, Han WY, et al. Seedling growth and seedling material accumulation of Polygonatum cyrtonema[J]. J Agric, 2021, 11(11): 76-80 | |
| [29] |
Li DD, Wang Q, Chen SS, et al. De novo assembly and analysis of Polygonatum cyrtonema Hua and identification of genes involved in polysaccharide and saponin biosynthesis[J]. BMC Genomics, 2022, 23(1): 195.
doi: 10.1186/s12864-022-08421-y |
| [30] |
Chen LS, Xu SW, Liu YJ, et al. Identification of key gene networks controlling polysaccharide accumulation in different tissues of Polygonatum cyrtonema Hua by integrating metabolic phenotypes and gene expression profiles[J]. Front Plant Sci, 2022, 13: 1012231.
doi: 10.3389/fpls.2022.1012231 URL |
| [31] | 单春苗, 王晨凯, 施圆圆, 等. 多花黄精甾体皂苷生物合成途径分析及关键酶基因研究[J]. 中国中药杂志, 2020, 45(12): 2847-2857. |
| Shan CM, Wang CK, Shi YY, et al. Identification of key enzyme genes involved in biosynthesis of steroidal saponins and analysis of biosynthesis pathway in Polygonatum cyrtonema[J]. China J Chin Mater Med, 2020, 45(12): 2847-2857. | |
| [32] | 廖荣俊, 杨阳, 叶碧欢, 等. 多花黄精根茎的转录组分析与甾体皂苷生物合成相关基因发掘[J]. 中国中药杂志, 2020, 45(7): 1648-1656. |
| Liao RJ, Yang Y, Ye BH, et al. Transcriptome analysis of rhizome of Polygonatum cyrtonema and identification of candidate genes involved in biosynthetic pathway of steroidal saponin[J]. China J Chin Mater Med, 2020, 45(7): 1648-1656. | |
| [33] |
Liang J, Chen Wd, Zong K, et al. Study on the interventional effects of Polygonatum cyrtonema polysaccharides on high-fat-diet-induced obese model mice through serum and liver metabolomics[J]. J Functional Foods, 2022, 95:105160.
doi: 10.1016/j.jff.2022.105160 URL |
| [1] | YANG Zhi-xiao, HOU Qian, LIU Guo-quan, LU Zhi-gang, CAO Yi, GOU Jian-yu, WANG Yi, LIN Ying-chao. Responses of Rubisco and Rubisco Activase in Different Resistant Tobacco Strains to Brown Spot Stress [J]. Biotechnology Bulletin, 2023, 39(9): 202-212. |
| [2] | YAO Sha-sha, WANG Jing-jing, WANG Jun-jie, LIANG Wei-hong. Molecular Mechanisms of Rice Grain Size Regulation Related to Plant Hormone Signaling Pathways [J]. Biotechnology Bulletin, 2023, 39(8): 80-90. |
| [3] | WANG Bing, ZHAO Hui-na, YU Jing, YU Shi-zhou, LEI Bo. Research Progress in the Regulation of Plant Branch Development [J]. Biotechnology Bulletin, 2023, 39(5): 14-22. |
| [4] | WANG Qi, HU Zhe, FU Wei, LI Guang-zhe, HAO Lin. Regulation of Burkholderia sp. GD17 on the Drought Tolerance of Cucumber Seedlings [J]. Biotechnology Bulletin, 2023, 39(3): 163-175. |
| [5] | YU Shi-xia, JIANG Yu-tong, LIN Wen-hui. Research Progress in Signals and Molecular Mechanisms of Ovule Primordia Initiation [J]. Biotechnology Bulletin, 2023, 39(2): 1-9. |
| [6] | SUN Yu-tong, LIU De-shuai, QI Xun, FENG Mei, HUANG Xu-zheng, YAO Wen-kong. Advances in Jasmonic Acid Regulating Plant Growth and Development as Well as Stress [J]. Biotechnology Bulletin, 2023, 39(11): 99-109. |
| [7] | YAN Meng-yu, WEI Xiao-wei, CAO Jing, LAN Hai-yan. Cloning of Basic Helix-loop-helix(bHLH)Transcription Factor Gene SabHLH169 in Suaeda aralocaspica and Analysis of Its Resistances to Drought Stress [J]. Biotechnology Bulletin, 2023, 39(11): 328-339. |
| [8] | RUAN Hang, DUO Hao-yuan, FAN Wen-yan, LV Qing-han, JIANG Shu-jun, ZHU Sheng-wei. Role of the AtERF49 in the Responses to Salt-alkali Stress in Arabidopsis [J]. Biotechnology Bulletin, 2023, 39(1): 150-156. |
| [9] | WEI Xin-xin, LAN Hai-yan. Advances in the Regulation of Plant MYB Transcription Factors in Secondary Metabolism and Stress Response [J]. Biotechnology Bulletin, 2022, 38(8): 12-23. |
| [10] | HONG Tian-shu, HAI Ying, ENHE Ba-ya-er, GAO Feng. Analysis of Expression Characteristics of CmABCG8 Gene in Cucumis melo L. [J]. Biotechnology Bulletin, 2022, 38(7): 178-185. |
| [11] | ZU Guo-qiang, HU Zhe, WANG Qi, LI Guang-zhe, HAO Lin. Regulatory Role of Burkholderia sp. GD17 in Rice Seedling’s Responses to Cadmium Stress [J]. Biotechnology Bulletin, 2022, 38(4): 153-162. |
| [12] | LI Bing-juan, ZHENG Lu, SHEN Ren-fang, LAN Ping. Proteomic Analysis of RPP1A Involved in the Seedling Growth of Arabidopsis thaliana [J]. Biotechnology Bulletin, 2022, 38(2): 10-20. |
| [13] | DANG Yuan, LI Wei, MIAO Xiang, XIU Yu, LIN Shan-zhi. Cloning of Oleosin Gene PsOLE4 from Prunus sibirica and Its Regulatory Function Analysis for Oil Accumulation [J]. Biotechnology Bulletin, 2022, 38(11): 151-161. |
| [14] | YIN Guo-liang, SUN Wen-hao, PANG Xiao-yun, SUN Fei. Application of cryo-Electron Microscopy in Molecular Botany Research [J]. Biotechnology Bulletin, 2022, 38(1): 15-32. |
| [15] | LI Wen-jiao, ZHANG Zhong-feng, LIU Qing, SUN Jie, YANG Li, WANG Xing-jun, ZHAO Shu-zhen. Role of BRs in Plant Response to Abiotic Stress [J]. Biotechnology Bulletin, 2022, 38(1): 228-235. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||