








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (12): 182-192.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0401
Previous Articles Next Articles
LIANG Jia-lin1( ), ZHAO Shuang1, LI Xing-er1, ZHAO Cheng-zhou2, LI Ping1(
), ZHAO Shuang1, LI Xing-er1, ZHAO Cheng-zhou2, LI Ping1( )
)
Received:2024-04-26
Online:2024-12-26
Published:2025-01-15
Contact:
LI Ping
E-mail:2455352154@qq.com;liping2015@qhu.edu.cn
LIANG Jia-lin, ZHAO Shuang, LI Xing-er, ZHAO Cheng-zhou, LI Ping. Identification of Aux/IAA Gene Family in Corydalis hendersonii Hemsl. and Analysis on Their Expression Pattern under UVB Treatment[J]. Biotechnology Bulletin, 2024, 40(12): 182-192.
| 引物名称Primer name | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| ChIAA1 | GCGAGCGAGATTCTGAGTGAGAG | GCATCCAGTCCCCATCCTTGTC |
| ChIAA2 | GATGTGCCGTGGGAGATGTTTATTG | TGGTGAGTGACTGGAGACAAGTTC |
| ChIAA3 | AGGATGCTGGTTGGAGATGATGTC | GTGTCATTGCTTGCTTGCTGTTTC |
| ChIAA4 | GATGGGTTGCGTGAGGAACAAAG | TGATGAGGGAAACAAAGAGGAAGGG |
| ChIAA5 | GATGGGTTGCGTGAGGAACAAAG | TGATGAGGGAAACAAAGAGGAAGGG |
| ChIAA6 | TAGAGGTGGAAATGGTTGTGGGAAG | CGCCTTGGGAGACATAGGAGTAAC |
| ChIAA7 | ATGCTATGAAACCCTTGCCCAAAC | AACCACATGCTCCTCTCCACTTG |
| ChIAA8 | GCGAGCGAGATTCTGAGTGAGAG | GCATCCAGTCCCCATCCTTGTC |
| ChIAA9 | TAGAGGTGGAAATGGTTGTGGGAAG | CGCCTTGGGAGACATAGGAGTAAC |
| ChIAA10 | AGGATGCTGGTTGGAGATGATGTC | GTGTCATTGCTTGCTTGCTGTTTC |
| ChIAA11 | CCATCGGCATCCAAAGCACAAG | CCTTCTCCTCATCGGCGTTACTC |
| ChIAA12 | AAGGAGACAGGATGTTGGTTGGAG | TGTCTTTCCTTGCTTGTTGCTTCC |
| ChIAA13 | GGAAGGAAGGAAGGAAGGAAGGAAG | AGTTGTAGTCGTCTTTGTCGTCTCC |
| ChIAA14 | GGAAGGAAGGAAGGAAGGAAGGAAG | AGTTGTAGTCGTCTTTGTCGTCTCC |
| ChIAA16 | AAGGAGACAGGATGTTGGTTGGAG | TGTCTTTCCTTGCTTGTTGCTTCC |
| ChIAA18 | GATGGGTTGCGTGAGGAACAAAG | TGATGAGGGAAACAAAGAGGAAGGG |
| ChIAA19 | TACGACGACGAAGACGAAGAACTC | CAAATCTTTCCTCCTCCGTTTTCCG |
| ChIAA21 | TAGAGGTGGAAATGGTTGTGGGAAG | CGCCTTGGGAGACATAGGAGTAAC |
| ChIAA23 | AGGATGCTGGTTGGAGATGATGTC | GTGTCATTGCTTGCTTGCTGTTTC |
| ChIAA24 | TTCTGATGCTTGTGACGATGTTGAC | GTTCTAACCGGCGGCCATCC |
| GAPDH | AGTGGACTTAACTTGCCGACTTGAG | GAACTGAGACCTATGCCAGCCTTG |
Table 1 Primer sequences used in this study
| 引物名称Primer name | 上游引物Forward primer(5'-3') | 下游引物Reverse primer(5'-3') |
|---|---|---|
| ChIAA1 | GCGAGCGAGATTCTGAGTGAGAG | GCATCCAGTCCCCATCCTTGTC |
| ChIAA2 | GATGTGCCGTGGGAGATGTTTATTG | TGGTGAGTGACTGGAGACAAGTTC |
| ChIAA3 | AGGATGCTGGTTGGAGATGATGTC | GTGTCATTGCTTGCTTGCTGTTTC |
| ChIAA4 | GATGGGTTGCGTGAGGAACAAAG | TGATGAGGGAAACAAAGAGGAAGGG |
| ChIAA5 | GATGGGTTGCGTGAGGAACAAAG | TGATGAGGGAAACAAAGAGGAAGGG |
| ChIAA6 | TAGAGGTGGAAATGGTTGTGGGAAG | CGCCTTGGGAGACATAGGAGTAAC |
| ChIAA7 | ATGCTATGAAACCCTTGCCCAAAC | AACCACATGCTCCTCTCCACTTG |
| ChIAA8 | GCGAGCGAGATTCTGAGTGAGAG | GCATCCAGTCCCCATCCTTGTC |
| ChIAA9 | TAGAGGTGGAAATGGTTGTGGGAAG | CGCCTTGGGAGACATAGGAGTAAC |
| ChIAA10 | AGGATGCTGGTTGGAGATGATGTC | GTGTCATTGCTTGCTTGCTGTTTC |
| ChIAA11 | CCATCGGCATCCAAAGCACAAG | CCTTCTCCTCATCGGCGTTACTC |
| ChIAA12 | AAGGAGACAGGATGTTGGTTGGAG | TGTCTTTCCTTGCTTGTTGCTTCC |
| ChIAA13 | GGAAGGAAGGAAGGAAGGAAGGAAG | AGTTGTAGTCGTCTTTGTCGTCTCC |
| ChIAA14 | GGAAGGAAGGAAGGAAGGAAGGAAG | AGTTGTAGTCGTCTTTGTCGTCTCC |
| ChIAA16 | AAGGAGACAGGATGTTGGTTGGAG | TGTCTTTCCTTGCTTGTTGCTTCC |
| ChIAA18 | GATGGGTTGCGTGAGGAACAAAG | TGATGAGGGAAACAAAGAGGAAGGG |
| ChIAA19 | TACGACGACGAAGACGAAGAACTC | CAAATCTTTCCTCCTCCGTTTTCCG |
| ChIAA21 | TAGAGGTGGAAATGGTTGTGGGAAG | CGCCTTGGGAGACATAGGAGTAAC |
| ChIAA23 | AGGATGCTGGTTGGAGATGATGTC | GTGTCATTGCTTGCTTGCTGTTTC |
| ChIAA24 | TTCTGATGCTTGTGACGATGTTGAC | GTTCTAACCGGCGGCCATCC |
| GAPDH | AGTGGACTTAACTTGCCGACTTGAG | GAACTGAGACCTATGCCAGCCTTG |
| 蛋白名称Protein name | α-螺旋α-helix/% | β-折叠β-angle/% | 随机卷曲Random coil/% | 延伸连 Extended strand/% |
|---|---|---|---|---|
| ChIAA1 | 19.85 | 3.31 | 59.03 | 17.81 |
| ChIAA2 | 26.23 | 6.49 | 46.23 | 21.04 |
| ChIAA3 | 18.67 | 4.8 | 61.87 | 14.67 |
| ChIAA4 | 16.02 | 4.14 | 64.64 | 15.19 |
| ChIAA5 | 18.01 | 2.77 | 63.71 | 15.51 |
| ChIAA6 | 13.75 | 3.44 | 66.76 | 16.05 |
| ChIAA7 | 22.84 | 5.25 | 53.09 | 18.83 |
| ChIAA8 | 16.09 | 2.21 | 65.93 | 15.77 |
| ChIAA9 | 18.59 | 2.56 | 61.86 | 16.99 |
| ChIAA10 | 19.80 | 2.97 | 61.39 | 15.84 |
| ChIAA11 | 23.21 | 3.07 | 52.22 | 21.50 |
| ChIAA12 | 17.29 | 3.01 | 56.77 | 22.93 |
| ChIAA13 | 26.52 | 3.03 | 55.68 | 14.77 |
| ChIAA14 | 22.96 | 3.89 | 57.20 | 15.95 |
| ChIAA16 | 17.97 | 3.69 | 56.68 | 21.66 |
| ChIAA18 | 7.73 | 2.42 | 76.81 | 13.04 |
| ChIAA19 | 20.77 | 3.86 | 58.45 | 16.91 |
| ChIAA21 | 31.89 | 4.86 | 47.03 | 16.22 |
| ChIAA23 | 10.30 | 1.21 | 70.91 | 17.58 |
| ChIAA24 | 26.14 | 5.88 | 45.75 | 22.22 |
Table 2 Secondary structure analysis of ChAux/IAA protein
| 蛋白名称Protein name | α-螺旋α-helix/% | β-折叠β-angle/% | 随机卷曲Random coil/% | 延伸连 Extended strand/% |
|---|---|---|---|---|
| ChIAA1 | 19.85 | 3.31 | 59.03 | 17.81 |
| ChIAA2 | 26.23 | 6.49 | 46.23 | 21.04 |
| ChIAA3 | 18.67 | 4.8 | 61.87 | 14.67 |
| ChIAA4 | 16.02 | 4.14 | 64.64 | 15.19 |
| ChIAA5 | 18.01 | 2.77 | 63.71 | 15.51 |
| ChIAA6 | 13.75 | 3.44 | 66.76 | 16.05 |
| ChIAA7 | 22.84 | 5.25 | 53.09 | 18.83 |
| ChIAA8 | 16.09 | 2.21 | 65.93 | 15.77 |
| ChIAA9 | 18.59 | 2.56 | 61.86 | 16.99 |
| ChIAA10 | 19.80 | 2.97 | 61.39 | 15.84 |
| ChIAA11 | 23.21 | 3.07 | 52.22 | 21.50 |
| ChIAA12 | 17.29 | 3.01 | 56.77 | 22.93 |
| ChIAA13 | 26.52 | 3.03 | 55.68 | 14.77 |
| ChIAA14 | 22.96 | 3.89 | 57.20 | 15.95 |
| ChIAA16 | 17.97 | 3.69 | 56.68 | 21.66 |
| ChIAA18 | 7.73 | 2.42 | 76.81 | 13.04 |
| ChIAA19 | 20.77 | 3.86 | 58.45 | 16.91 |
| ChIAA21 | 31.89 | 4.86 | 47.03 | 16.22 |
| ChIAA23 | 10.30 | 1.21 | 70.91 | 17.58 |
| ChIAA24 | 26.14 | 5.88 | 45.75 | 22.22 |
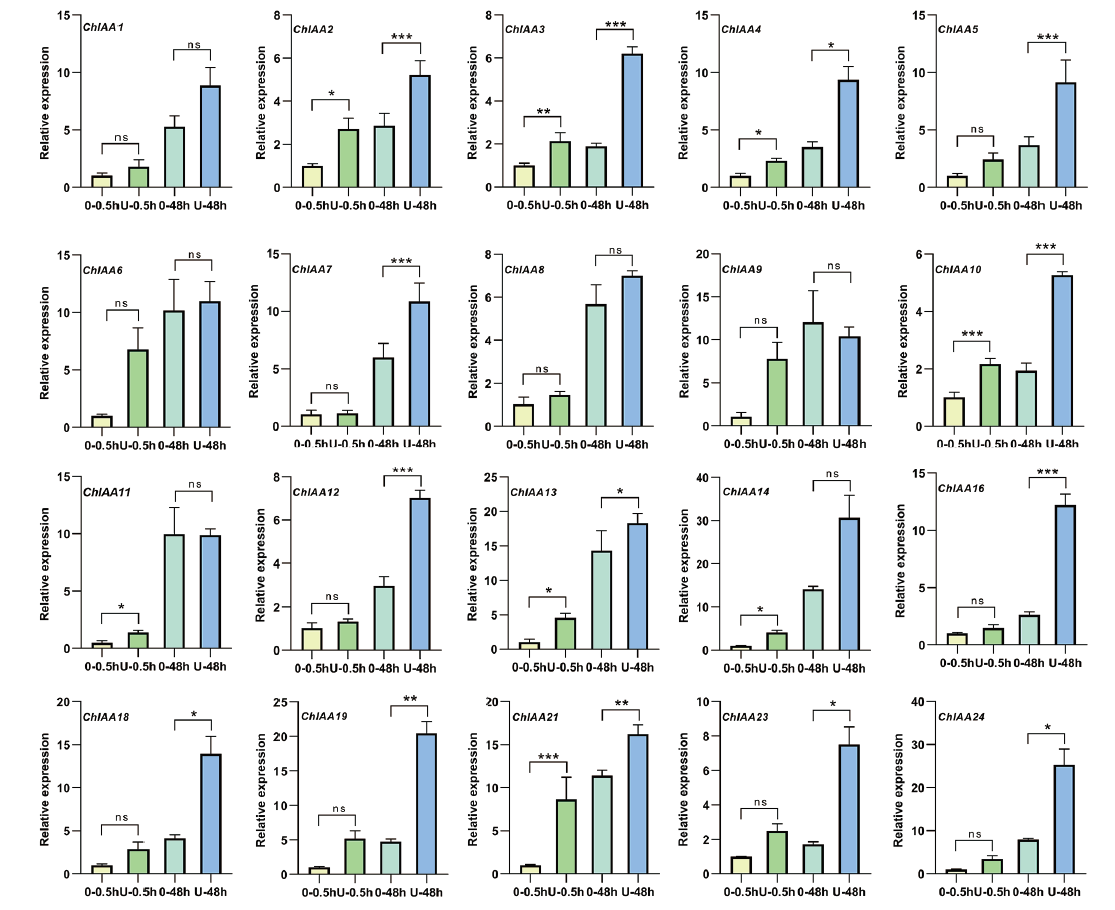
Fig. 7 Relative expressions of ChAux/IAA genes under UVB treatment conditions 0-0.5h indicates untreated 0.5 h;U-0.5h indicates UVB treated for 0.5 h; 0-48h indicates untreated 48 h;U-48h indicates UVB treated for 48 h. *P < 0.05, **P < 0.01, ***P < 0.001
| [1] | 中国植物志编辑委员会. 中国植物志·第40卷[M]. 北京: 科学出版社,1998: 80. |
| Editorial Committee of Flora of China. Flora of China, Volume 40[M]. Beijing: Science Press, 1998: 80. | |
| [2] | 李雪, 马海梅, 任玉玲, 等. 尼泊尔黄堇G6PD基因家族的鉴定及生物信息学分析[J/OL]. 分子植物育种, 2023. https://kns.cnki.net/kcms/detail/46.1068.S.20230314.1825.029.html. |
| Li X, Ma HM, Ren YL, et al. Identification and bioinformatics analysis of G6PD gene family of Corydalis hendersonii hensel[J/OL]. Mol Plant Breed, 2023. https://kns.cnki.net/kcms/detail/46.1068.S.20230314.1825.029.html. | |
| [3] | 魏春华, 程虹毓, 高燕萍, 等. 藏药矮紫堇解热镇痛抗炎作用的研究[J]. 中国新药杂志, 2017, 26(3): 337-342. |
| Wei CH, Cheng HY, Gao YP, et al. Antipyretic, analgesic and anti-inflammatory effects of Corydalis hendersonii Hemsl[J]. Chin J N Drugs, 2017, 26(3): 337-342. | |
| [4] | Jin JH, Zhang HX, Tan JY, et al. A new ethylene-responsive factor CaPTI1 gene of pepper(Capsicum annuum L.) involved in the regulation of defense response to Phytophthora capsici[J]. Front Plant Sci, 2016, 6: 1217. |
| [5] | 王国平, 周东洁, 牛永志, 等. 普通烟草Aux/IAA转录因子家族全基因组鉴定分析[J]. 中国烟草学报, 2019, 25(6): 80-89. |
| Wang GP, Zhou DJ, Niu YZ, et al. Genome-wide identification and analysis of Aux/IAA gene family in Nicotiana tobacum[J]. Acta Tabacaria Sin, 2019, 25(6): 80-89. | |
| [6] | Wang SK, Bai YH, Shen CJ, et al. Auxin-related gene families in abiotic stress response in Sorghum bicolor[J]. Funct Integr Genom, 2010, 10(4): 533-546. |
| [7] | Çakir B, Kiliçkaya O, Olcay AC. Genome-wide analysis of Aux/IAA genes in Vitis vinifera: cloning and expression profiling of a grape Aux/IAA gene in response to phytohormone and abiotic stresses[J]. Acta Physiol Plant, 2013, 35(2): 365-377. |
| [8] | Farcot E, Lavedrine C, Vernoux T. A modular analysis of the auxin signalling network[J]. PLoS One, 2015, 10(3): e0122231. |
| [9] |
Hu W, Zuo J, Hou XW, et al. The auxin response factor gene family in banana: genome-wide identification and expression analyses during development, ripening, and abiotic stress[J]. Front Plant Sci, 2015, 6: 742.
doi: 10.3389/fpls.2015.00742 pmid: 26442055 |
| [10] |
Overvoorde PJ, Okushima Y, Alonso JM, et al. Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana[J]. Plant Cell, 2005, 17(12): 3282-3300.
doi: 10.1105/tpc.105.036723 pmid: 16284307 |
| [11] | Liscum E, Reed JW. Genetics of Aux/IAA and ARF action in plant growth and development[J]. Plant Mol Biol, 2002, 49(3/4): 387-400. |
| [12] | Luo J, Zhou JJ, Zhang JZ. Aux/IAA gene family in plants: molecular structure, regulation, and function[J]. Int J Mol Sci, 2018, 19(1): 259. |
| [13] | Salehin M, Li BH, Tang M, et al. Auxin-sensitive Aux/IAA proteins mediate drought tolerance in Arabidopsis by regulating glucosinolate levels[J]. Nat Commun, 2019, 10(1): 4021. |
| [14] | Mielecki J, Gawroński P, Karpiński S. Aux/IAA11 is required for UV-AB tolerance and auxin sensing in Arabidopsis thaliana[J]. Int J Mol Sci, 2022, 23(21): 13386. |
| [15] |
Nakamura A, Umemura I, Gomi K, et al. Production and characterization of auxin-insensitive rice by overexpression of a mutagenized rice IAA protein[J]. Plant J, 2006, 46(2): 297-306.
doi: 10.1111/j.1365-313X.2006.02693.x pmid: 16623891 |
| [16] | Ni J, Wang GH, Zhu ZX, et al. OsIAA23-mediated auxin signaling defines postembryonic maintenance of QC in rice[J]. Plant J, 2011, 68(3): 433-442. |
| [17] | Zhu ZX, Liu Y, Liu SJ, et al. A gain-of-function mutation in OsIAA11 affects lateral root development in rice[J]. Mol Plant, 2012, 5(1): 154-161. |
| [18] |
Jain M, Khurana JP. Transcript profiling reveals diverse roles of auxin-responsive genes during reproductive development and abiotic stress in rice[J]. FEBS J, 2009, 276(11): 3148-3162.
doi: 10.1111/j.1742-4658.2009.07033.x pmid: 19490115 |
| [19] |
Jung H, Lee DK, Choi YD, et al. OsIAA6, a member of the rice Aux/IAA gene family, is involved in drought tolerance and tiller outgrowth[J]. Plant Sci, 2015, 236: 304-312.
doi: 10.1016/j.plantsci.2015.04.018 pmid: 26025543 |
| [20] | Zhang AY, Yang X, Lu J, et al. OsIAA20, an Aux/IAA protein, mediates abiotic stress tolerance in rice through an ABA pathway[J]. Plant Sci, 2021, 308: 110903. |
| [21] |
Song YL, You J, Xiong LZ. Characterization of OsIAA1 gene, a member of rice Aux/IAA family involved in auxin and brassinosteroid hormone responses and plant morphogenesis[J]. Plant Mol Biol, 2009, 70(3): 297-309.
doi: 10.1007/s11103-009-9474-1 pmid: 19266169 |
| [22] | Song YL, Xu ZF. Ectopic overexpression of an AUXIN/INDOLE-3-ACETIC ACID(Aux/IAA)gene OsIAA4 in rice induces morphological changes and reduces responsiveness to auxin[J]. Int J Mol Sci, 2013, 14(7): 13645-13656. |
| [23] | Zhang SN, Wang SK, Xu YX, et al. The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1[J]. Plant Cell Environ, 2015, 38(4): 638-654. |
| [24] | Chen SH, Zhou LJ, Xu P, et al. SPOC domain-containing protein leaf inclination3 interacts with LIP1 to regulate rice leaf inclination through auxin signaling[J]. PLoS Genet, 2018, 14(11): e1007829. |
| [25] | 张战营. 水稻产量基因Gnp4的克隆与功能机理分析[D]. 北京: 中国农业大学, 2015. |
| Zhang ZY. Cloning and functional mechanism analysis of rice yield gene Gnp4[D]. Beijing: China Agricultural University, 2015. | |
| [26] | Kim GE, Kim MS, Sung J. UVB irradiation-induced transcriptional changes in lignin- and flavonoid biosynthesis and indole/tryptophan-auxin-responsive genes in rice seedlings[J]. Plants, 2022, 11(12): 1618. |
| [27] | Singla B, Chugh A, Khurana JP, et al. An early auxin-responsive Aux/IAA gene from wheat(Triticum aestivum)is induced by epibrassinolide and differentially regulated by light and calcium[J]. J Exp Bot, 2006, 57(15): 4059-4070. |
| [28] |
Chen DD, Richardson T, Chai SC, et al. Drought-up-regulated TaNAC69-1 is a transcriptional repressor of TaSHY2 and TaIAA7, and enhances root length and biomass in wheat[J]. Plant Cell Physiol, 2016, 57(10): 2076-2090.
pmid: 27440550 |
| [29] | Jia ML, Li YN, Wang ZY, et al. TaIAA21 represses TaARF25-mediated expression of TaERFs required for grain size and weight development in wheat[J]. Plant J, 2021, 108(6): 1754-1767. |
| [30] |
Bassa C, Mila I, Bouzayen M, et al. Phenotypes associated with down-regulation of Sl-IAA27 support functional diversity among Aux/IAA family members in tomato[J]. Plant Cell Physiol, 2012, 53(9): 1583-1595.
doi: 10.1093/pcp/pcs101 pmid: 22764281 |
| [31] |
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets[J]. Mol Biol Evol, 2016, 33(7): 1870-1874.
doi: 10.1093/molbev/msw054 pmid: 27004904 |
| [32] |
Chen CJ, Chen H, Zhang Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant, 2020, 13(8): 1194-1202.
doi: S1674-2052(20)30187-8 pmid: 32585190 |
| [33] |
李艳艳, 齐艳华. 植物Aux/IAA基因家族生物学功能研究进展[J]. 植物学报, 2022, 57(1): 30-41.
doi: 10.11983/CBB21168 |
| Li YY, Qi YH. Advances in biological functions of Aux/IAA gene family in plants[J]. Chin Bull Bot, 2022, 57(1): 30-41. | |
| [34] | 苏丽艳. 猕猴桃Aux/IAA基因家族的鉴定与表达[J]. 西北植物学报, 2023, 43(1): 55-65. |
| Su LY. Identification and expression analysis of Aux/IAA gene family in Actinidia chinensis[J]. Acta Bot Boreali Occidentalia Sin, 2023, 43(1): 55-65. | |
| [35] |
Reed JW. Roles and activities of Aux/IAA proteins in Arabidopsis[J]. Trends Plant Sci, 2001, 6(9): 420-425.
doi: 10.1016/s1360-1385(01)02042-8 pmid: 11544131 |
| [36] | Yang XF, Gao SH, Guo L, et al. Three chromosome-scale Papaver genomes reveal punctuated patchwork evolution of the morphinan and noscapine biosynthesis pathway[J]. Nat Commun, 2021, 12(1): 6030. |
| [37] | 冯曦, 豆浩, 孙佳佳, 等. 桑树Aux/IAA基因家族鉴定与IBA处理下表达模式分析[J]. 河南农业大学学报, 2023, 57(6): 936-948. |
| Feng X, Dou H, Sun JJ, et al. Identification of Aux/IAA gene family of Morus notabilis and analysis of expression patterns under IBA treatment[J]. J Henan Agric Univ, 2023, 57(6): 936-948. | |
| [38] | 刘湘楠, 胡祥祥, 乔梦燚, 等. 红花Aux/IAA基因家族鉴定及盐胁迫响应分析[J]. 江苏农业科学, 2024, 52(6): 68-76. |
| Liu XN, Hu XX, Qiao MY, et al. Identification of Aux/IAA gene family of Carthamus tinctorius and analysis of salt stress response[J]. Jiangsu Agric Sci, 2024, 52(6): 68-76. | |
| [39] | 周光锋, 季霞, 罗磊, 等. 青檀(Pteroceltis tatarinowii)Aux/IAA基因的发掘及其在不同施氮水平下的表达[J/OL]. 分子植物育种, 2023. https://kns.cnki.net/kcms/detail/46.1068.S.20231120.1631.022.html. |
| Zhou GF, Ji X, Luo L, et al. Identification of Aux/IAA genes in Pteroceltis tatarinowii and their expression under different nitrogen fertilizer application levels[J/OL]. Mol Plant Breed, 2023. https://kns.cnki.net/kcms/detail/46.1068.S.20231120.1631.022.html. | |
| [40] | 冯昭, 刘世鹏, 吕蕊花, 等. 决明Aux/IAA和ARF基因家族鉴定、表达及蛋白互作分析[J]. 药学学报, 2024, 59(3): 751-763. |
| Feng Z, Liu SP, Lü RH, et al. Identification, expression and protein interaction analysis of Aux/IAA and ARF gene family in Senna tora L[J]. Acta Pharm Sin, 2024, 59(3): 751-763. | |
| [41] |
乔麟轶, 李欣, 畅志坚, 等. 粗山羊草全基因组 Aux/IAA 基因家族的分离、染色体定位及序列分析[J]. 作物学报, 2014, 40(12): 2059-2069.
doi: 10.3724/SP.J.1006.2014.02059 |
| Qiao LY, Li X, Chang ZJ, et al. Whole-genome sequence isolation, chromosome location, and characterization of primary auxin-responsive Aux/IAA gene family in Aegilops tauschii[J]. Acta Agron Sin, 2014, 40(12): 2059-2069. | |
| [42] | 王垒, 陈劲枫, 贾利. 黄瓜Aux/IAA基因家族的生物信息学分析[J]. 中国瓜菜, 2010, 23(6): 1-4. |
| Wang L, Chen JF, Jia L. Bioinformatic analysis of the Aux/IAA gene family in cucumber[J]. China Cucurbits Veg, 2010, 23(6): 1-4. | |
| [43] | 郑小敏, 赵敬会, 李荣冲, 等. 芸薹属大白菜Aux/IAA基因家族的生物信息学分析[J]. 北方园艺, 2012(14): 109-113. |
| Zheng XM, Zhao JH, Li RC, et al. Bioinformtics analysis of primary auxin-responsive Aux/IAA gene family in Brassica rapa[J]. North Hortic, 2012(14): 109-113. | |
| [44] |
王益军, 吕燕萍, 谢秦, 等. 高粱全基因组生长素原初响应基因Aux/IAA的序列特征分析[J]. 作物学报, 2010, 36(4): 688-694.
doi: 10.3724/SP.J.1006.2010.00688 |
| Wang YJ, Lv YP, Xie Q, et al. Whole-genome sequence characterization of primary auxin-responsive Aux/IAA gene family in sorghum(Sorghum bicolor L.)[J]. Acta Agron Sin, 2010, 36(4): 688-694. | |
| [45] | Liu SD, Hu QN, Luo S, et al. Expression of wild-type PtrIAA14.1, a poplar Aux/IAA gene causes morphological changes in Arabidop-sis[J]. Front Plant Sci, 2015, 6: 388. |
| [46] |
Yu H, Soler M, San Clemente H, et al. Comprehensive genome-wide analysis of the Aux/IAA gene family in Eucalyptus: evidence for the role of EgrIAA4 in wood formation[J]. Plant Cell Physiol, 2015, 56(4): 700-714.
doi: 10.1093/pcp/pcu215 pmid: 25577568 |
| [47] |
Yamauchi T, Tanaka A, Inahashi H, et al. Fine control of aerenchyma and lateral root development through AUX/IAA- and ARF-dependent auxin signaling[J]. Proc Natl Acad Sci USA, 2019, 116(41): 20770-20775.
doi: 10.1073/pnas.1907181116 pmid: 31548376 |
| [48] | 张佩佩. MfGATA25、MfIDD1和MtIAA15调控苜蓿生长与耐寒性的功能研究[D]. 广州: 华南农业大学, 2019. |
| Zhang PP. Study on the function of MfGATA25, MfIDD1 and MTIAAA 15 in regulating alfalfa growth and cold tolerance[D]. Guangzhou: South China Agricultural University, 2019. | |
| [49] | Findlay KMW, Jenkins GI. Regulation of UVR8 photoreceptor dimer/monomer photo-equilibrium in Arabidopsis plants grown under photoperiodic conditions[J]. Plant Cell Environ, 2016, 39(8): 1706-1714. |
| [50] |
Yadav A, Singh D, Lingwan M, et al. Light signaling and UV-B-mediated plant growth regulation[J]. J Integr Plant Biol, 2020, 62(9): 1270-1292.
doi: 10.1111/jipb.12932 |
| [51] | Jansen MAK. Ultraviolet-B radiation effects on plants: induction of morphogenic responses[J]. Physiol Plant, 2002, 116(3): 423-429. |
| [52] | Favory JJ, Stec A, Gruber H, et al. Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis[J]. EMBO J, 2009, 28(5): 591-601. |
| [53] |
Vanhaelewyn L, Prinsen E, Van Der Straeten D, et al. Hormone-controlled UV-B responses in plants[J]. J Exp Bot, 2016, 67(15): 4469-4482.
doi: 10.1093/jxb/erw261 pmid: 27401912 |
| [54] |
Jansen MAK, Coffey AM, Prinsen E. UV-B induced morphogenesis: four players or a quartet?[J]. Plant Signal Behav, 2012, 7(9): 1185-1187.
doi: 10.4161/psb.21260 pmid: 22899069 |
| [55] |
Qian CZ, Chen ZR, Liu Q, et al. Coordinated transcriptional regulation by the UV-B photoreceptor and multiple transcription factors for plant UV-B responses[J]. Mol Plant, 2020, 13(5): 777-792.
doi: S1674-2052(20)30062-9 pmid: 32126287 |
| [56] | Liu XM, Sun WY, Ma B, et al. Genome-wide analysis of blueberry B-box family genes and identification of members activated by abiotic stress[J]. BMC Genomics, 2023, 24(1): 584. |
| [57] |
Jiang L, Wang Y, Li QF, et al. Arabidopsis STO/BBX24 negatively regulates UV-B signaling by interacting with COP1 and repressing HY5 transcriptional activity[J]. Cell Res, 2012, 22(6): 1046-1057.
doi: 10.1038/cr.2012.34 pmid: 22410790 |
| [58] |
Fang HC, Dong YH, Yue XX, et al. MdCOL4 interaction mediates crosstalk between UV-B and high temperature to control fruit coloration in apple[J]. Plant Cell Physiol, 2019, 60(5): 1055-1066.
doi: 10.1093/pcp/pcz023 pmid: 30715487 |
| [59] | Liu X, Li R, Dai YQ, et al. Genome-wide identification and expression analysis of the B-box gene family in the apple(Malus domestica Borkh.) genome[J]. Mol Genet Genomics, 2018, 293(2): 303-315. |
| [60] |
Kim J, Harter K, Theologis A. Protein-protein interactions among the Aux/IAA proteins[J]. Proc Natl Acad Sci USA, 1997, 94(22): 11786-11791.
doi: 10.1073/pnas.94.22.11786 pmid: 9342315 |
| [1] | DU Pin-ting, WU Guo-jiang, WANG Zhen-guo, LI Yan, ZHOU Wei, ZHOU Ya-xing. Identification and Expression Analysis of CPP Gene Family in Sorghum [J]. Biotechnology Bulletin, 2025, 41(1): 132-142. |
| [2] | WANG Zi-ao, TIAN Rui, CUI Yong-mei, BAI Yi-xiong, YAO Xiao-hua, AN Li-kun, WU Kun-lun. Bioinformatics and Expression Pattern Analysis of HvnJAZ4 Gene in Hulless Barley [J]. Biotechnology Bulletin, 2025, 41(1): 173-185. |
| [3] | KONG Qing-yang, ZHANG Xiao-long, LI Na, ZHANG Chen-jie, ZHANG Xue-yun, YU Chao, ZHANG Qi-xiang, LUO Le. Identification and Expression Analysis of GRAS Transcription Factor Family in Rosa persica [J]. Biotechnology Bulletin, 2025, 41(1): 210-220. |
| [4] | KOU Bei-sen, CHENG Meng-meng, GUO Xue-qin, GE Bin, LIU Di, LU Hai, LI Hui. Effects of Histone Deacetylase Inhibitor TSA Treatment on the Stem Development of Poplar [J]. Biotechnology Bulletin, 2025, 41(1): 240-251. |
| [5] | PEI Xu-juan, DI Jing-yi, LIU Hao, GAO Wei-xia. Exploration of Regulatory Elements for Hyaluronic Acid Molecular Weight in Streptococcus zooepidemicus via Transcriptome Analysis [J]. Biotechnology Bulletin, 2025, 41(1): 347-356. |
| [6] | WU Hui-qin, WANG Yan-hong, LIU Han, SI Zheng, LIU Xue-qing, WANG Jing, YANG Yi, CHENG Yan. Identification and Expression Analysis of UGT Gene Family in Pepper [J]. Biotechnology Bulletin, 2024, 40(9): 198-211. |
| [7] | TAN Bo-wen, ZHANG Yi, ZHANG Peng, WANG Zhen-yu, MA Qiu-xiang. Identification and Bioinformatics Analysis of Gene in the Magnesium Transporter Family in Cassava [J]. Biotechnology Bulletin, 2024, 40(9): 20-32. |
| [8] | MAN Quan-cai, MENG Zi-nuo, LI Wei, CAI Xin-ru, SU Run-dong, FU Chang-qing, GAO Shun-juan, CUI Jiang-hui. Identification and Expression Analysis of AQP Gene Family in Potato [J]. Biotechnology Bulletin, 2024, 40(9): 51-63. |
| [9] | WU Juan, WU Xiao-juan, WANG Pei-jie, XIE Rui, NIE Hu-shuai, LI Nan, MA Yan-hong. Screening and Expression Analysis of ERF Gene Related to Anthocyanin Synthesis in Colored Potato [J]. Biotechnology Bulletin, 2024, 40(9): 82-91. |
| [10] | SONG Bing-fang, LIU Ning, CHENG Xin-yan, XU Xiao-bin, TIAN Wen-mao, GAO Yue, BI Yang, WANG Yi. Identification of Potato G6PDH Gene Family and Its Expression Analysis in Damaged Tubers [J]. Biotechnology Bulletin, 2024, 40(9): 104-112. |
| [11] | WU Shuai, XIN Yan-ni, MAI Chun-hai, MU Xiao-ya, WANG Min, YUE Ai-qin, ZHAO Jin-zhong, WU Shen-jie, DU Wei-jun, WANG Li-xiang. Genome-wide Identification and Stress Response Analysis of Soybean GS Gene Family [J]. Biotechnology Bulletin, 2024, 40(8): 63-73. |
| [12] | NIE Zhu-xin, GUO Jin, QIAO Zi-yang, LI Wei-wei, ZHANG Xue-yan, LIU Chun-yang, WANG Jing. Transcriptome Analysis of the Anthocyanin Biosynthesis in the Fruit Development Processes of Lycium ruthenicum Murr. [J]. Biotechnology Bulletin, 2024, 40(8): 106-117. |
| [13] | YANG Wei, ZHAO Li-fen, TANG Bing, ZHOU Lin-bi, YANG Juan, MO Chuan-yuan, ZHANG Bao-hui, LI Fei, RUAN Song-lin, DENG Ying. Genome-wide Identification and Expression Analysis of the SRO Gene Family in Brassica juncea L. [J]. Biotechnology Bulletin, 2024, 40(8): 129-141. |
| [14] | ZHOU Lin, HUANG Shun-man, SU Wen-kun, YAO Xiang, QU Yan. Identification of the bHLH Gene Family and Selection of Genes Related to Color Formation in Camellia reticulata [J]. Biotechnology Bulletin, 2024, 40(8): 142-151. |
| [15] | GAO Meng-meng, ZHAO Tian-yu, JIAO Xin-yue, LIN Chun-jing, GUAN Zhe-yun, DING Xiao-yang, SUN Yan-yan, ZHANG Chun-bao. Comparative Transcriptome Analysis of Cytoplasmic Male Sterile Line and Its Restorer Line in Soybean [J]. Biotechnology Bulletin, 2024, 40(7): 137-149. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||