








Biotechnology Bulletin ›› 2024, Vol. 40 ›› Issue (9): 64-73.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0133
Previous Articles Next Articles
SHEN Peng( ), GAO Ya-Bin(
), GAO Ya-Bin( ), DING Hong
), DING Hong
Received:2024-02-02
Online:2024-09-26
Published:2024-06-26
Contact:
GAO Ya-Bin
E-mail:lzbwkjxy_sp@126.com;lzbwkjxy_gyb@126.com
SHEN Peng, GAO Ya-Bin, DING Hong. Identification and Expression Analysis of SAT Gene Family in Potato(Solanum tuberosum L.)[J]. Biotechnology Bulletin, 2024, 40(9): 64-73.
| 基因ID Gene ID | 正向引物Forward primer(5'-3') | 反向引物Reverse primer(5'-3') |
|---|---|---|
| Soltu06G005680 | GGTCGTGTTGAGACTGGTGTGATC | GCTTCGTGGTGCATCTCTACAGAC |
| Soltu02G022850 | CTGTCGCTAACACCGCTTCTACTC | GCTCAATGCCCGTTCCAAACAATC |
| Soltu12G025770 | GACACGGCGGAAGAAGCTATCTG | AGTGAGCGTTCAAGCGAAGAGTG |
| Soltu07G027060 | CCAAACAAGCCCCAAATCGACAAC | GCAGGAAGCAAGGTCAGGGAAAG |
| Soltu10G001180 | TGGTGCTGGTACTTGTGTTCTTGG | AGCAGTAGTTCTTGCAGGCACTTC |
Table 1 Primer information for qPCR
| 基因ID Gene ID | 正向引物Forward primer(5'-3') | 反向引物Reverse primer(5'-3') |
|---|---|---|
| Soltu06G005680 | GGTCGTGTTGAGACTGGTGTGATC | GCTTCGTGGTGCATCTCTACAGAC |
| Soltu02G022850 | CTGTCGCTAACACCGCTTCTACTC | GCTCAATGCCCGTTCCAAACAATC |
| Soltu12G025770 | GACACGGCGGAAGAAGCTATCTG | AGTGAGCGTTCAAGCGAAGAGTG |
| Soltu07G027060 | CCAAACAAGCCCCAAATCGACAAC | GCAGGAAGCAAGGTCAGGGAAAG |
| Soltu10G001180 | TGGTGCTGGTACTTGTGTTCTTGG | AGCAGTAGTTCTTGCAGGCACTTC |
| 基因ID Gene ID | 染色体定位 Chromosome localization | 氨基酸长度 Amino acid length | 相对分子量 Molecular weight/ kD | 等电点 Point isoelectric(pI) | ||
|---|---|---|---|---|---|---|
| Soltu.DM.02G022850 | chr02 | 36 554 173-36 560 967 | 349 | 37.43 | 6.41 | |
| Soltu.DM.07G027060 | chr07 | 56 287 590-56 289 231 | 359 | 39.04 | 8.80 | |
| Soltu.DM.10G001180 | chr10 | 978 595-980 158 | 365 | 40.23 | 6.50 | |
| Soltu.DM.12G025770 | chr12 | 55 741 243-55 744 365 | 293 | 31.27 | 6.38 | |
Table 2 Chromosomal localization and physicochemical properties of StSATs
| 基因ID Gene ID | 染色体定位 Chromosome localization | 氨基酸长度 Amino acid length | 相对分子量 Molecular weight/ kD | 等电点 Point isoelectric(pI) | ||
|---|---|---|---|---|---|---|
| Soltu.DM.02G022850 | chr02 | 36 554 173-36 560 967 | 349 | 37.43 | 6.41 | |
| Soltu.DM.07G027060 | chr07 | 56 287 590-56 289 231 | 359 | 39.04 | 8.80 | |
| Soltu.DM.10G001180 | chr10 | 978 595-980 158 | 365 | 40.23 | 6.50 | |
| Soltu.DM.12G025770 | chr12 | 55 741 243-55 744 365 | 293 | 31.27 | 6.38 | |
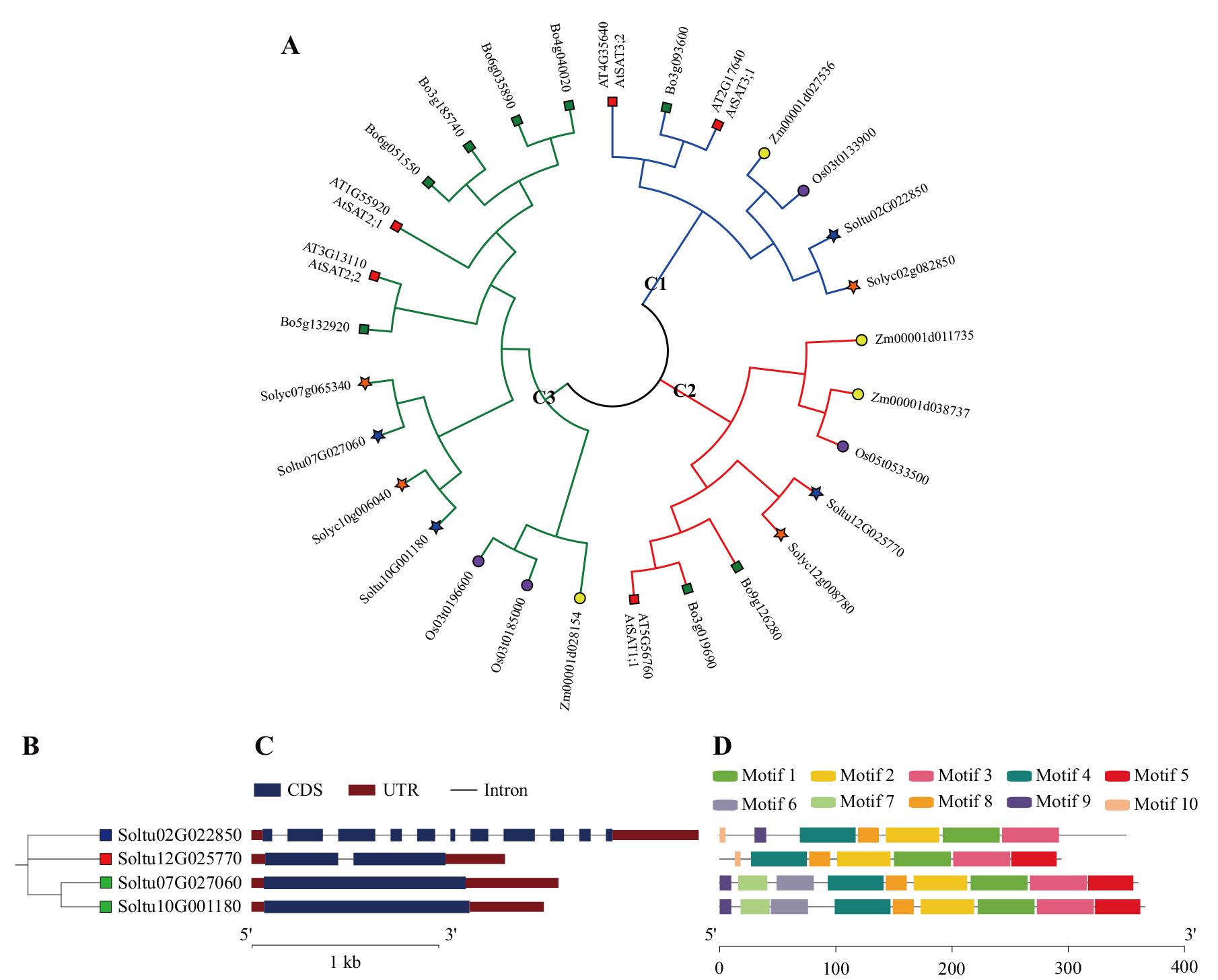
Fig. 1 Phylogenetic tree of SAT proteins in multiple species and analysis in evolutionary relationship, gene structure and protein conserved motifs of StSAT gene family members A: Phylogenetic tree of SAT proteins in multiple species. The different colored lines represent the three subfamilies(blue for C1, red for C2, and green for C3). The blue pentagrams indicate the potato SAT gene family member(StSATs), orange pentagrams indicate the tomato SAT gene family member(SlSATs), yellow circles indicate the corn SAT gene family member(ZmSATs), purple circles indicate the rice SAT gene family member(OsSATs), red squares indicate the Arabidopsis SAT gene family members(AtSTAs), and green squares indicate the cabbage SAT gene family member(BoSATs). B: Evolutionary tree of the StSAT gene family. The blue squares indicate the C1 subfamily, the red squares indicate the C2 subfamily, and the green squares indicate the C3 subfamily. C: Gene structure of StSATs. Blue boxes indicate exons, black lines indicate introns and red boxes indicate upstream/downstream regions. D: Protein conserved motif of StSATs
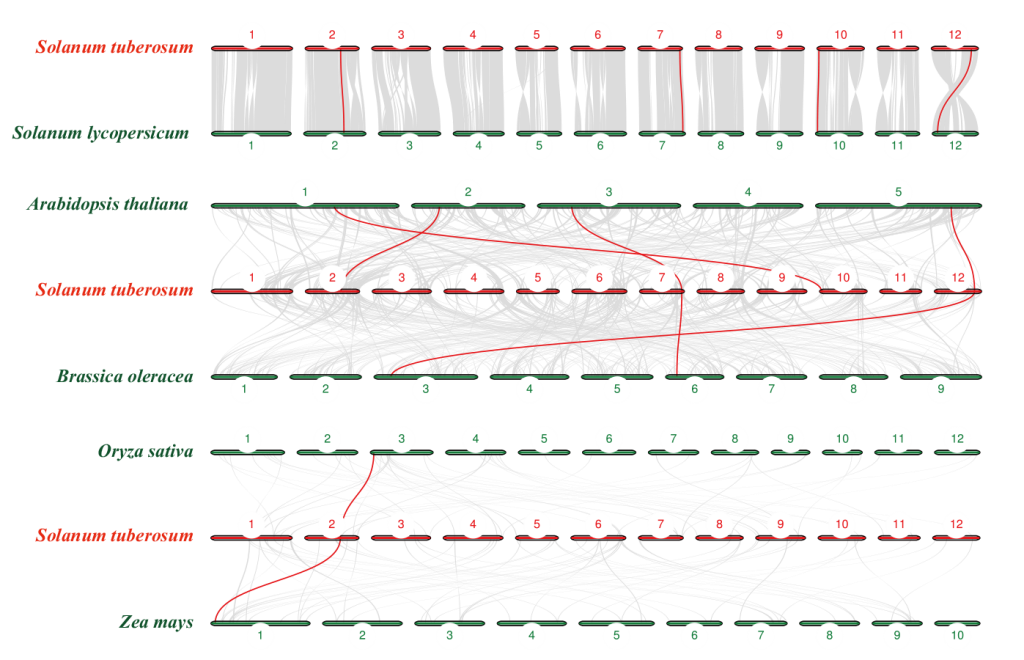
Fig. 2 Collinearity analysis of SAT genes in multiple species Different numerical numbers indicate the chromosomes of different species, and the red lines indicate orthologous gene pairs between different species
| Gene ID1 | Gene ID2 | Ka | Ks | Ka/Ks |
|---|---|---|---|---|
| Soltu02G022850 | Solyc02g082850 | 0.040 2 | 0.110 7 | 0.363 2 |
| Soltu12G025770 | Solyc12g008780 | 0.003 1 | 0.074 1 | 0.041 4 |
| Soltu07G027060 | Solyc07g065340 | 0.010 6 | 0.072 8 | 0.145 3 |
| Soltu10G001180 | Solyc10g006040 | 0.009 5 | 0.050 7 | 0.187 0 |
| Soltu02G022850 | AT2G17640 | 0.192 7 | 1.660 3 | 0.116 1 |
| Soltu12G025770 | AT5G56760 | 0.118 8 | 2.718 1 | 0.043 7 |
| Soltu07G027060 | AT3G13110 | 0.238 7 | 2.356 3 | 0.101 3 |
| Soltu10G001180 | AT1G55920 | 0.234 9 | 3.864 2 | 0.060 8 |
| Soltu12G025770 | Bo3g019690 | 0.122 7 | 3.050 0 | 0.040 2 |
| Soltu07G027060 | Bo6g035890 | 0.202 6 | 3.867 2 | 0.052 4 |
| Soltu02G022850 | Zm00001d027536 | 0.178 5 | 2.483 9 | 0.071 8 |
| Soltu02G022850 | Os03g04140 | 0.267 1 | 3.167 0 | 0.084 3 |
Table 3 Ka/Ks ratios of orthologous genes between potato and other species
| Gene ID1 | Gene ID2 | Ka | Ks | Ka/Ks |
|---|---|---|---|---|
| Soltu02G022850 | Solyc02g082850 | 0.040 2 | 0.110 7 | 0.363 2 |
| Soltu12G025770 | Solyc12g008780 | 0.003 1 | 0.074 1 | 0.041 4 |
| Soltu07G027060 | Solyc07g065340 | 0.010 6 | 0.072 8 | 0.145 3 |
| Soltu10G001180 | Solyc10g006040 | 0.009 5 | 0.050 7 | 0.187 0 |
| Soltu02G022850 | AT2G17640 | 0.192 7 | 1.660 3 | 0.116 1 |
| Soltu12G025770 | AT5G56760 | 0.118 8 | 2.718 1 | 0.043 7 |
| Soltu07G027060 | AT3G13110 | 0.238 7 | 2.356 3 | 0.101 3 |
| Soltu10G001180 | AT1G55920 | 0.234 9 | 3.864 2 | 0.060 8 |
| Soltu12G025770 | Bo3g019690 | 0.122 7 | 3.050 0 | 0.040 2 |
| Soltu07G027060 | Bo6g035890 | 0.202 6 | 3.867 2 | 0.052 4 |
| Soltu02G022850 | Zm00001d027536 | 0.178 5 | 2.483 9 | 0.071 8 |
| Soltu02G022850 | Os03g04140 | 0.267 1 | 3.167 0 | 0.084 3 |
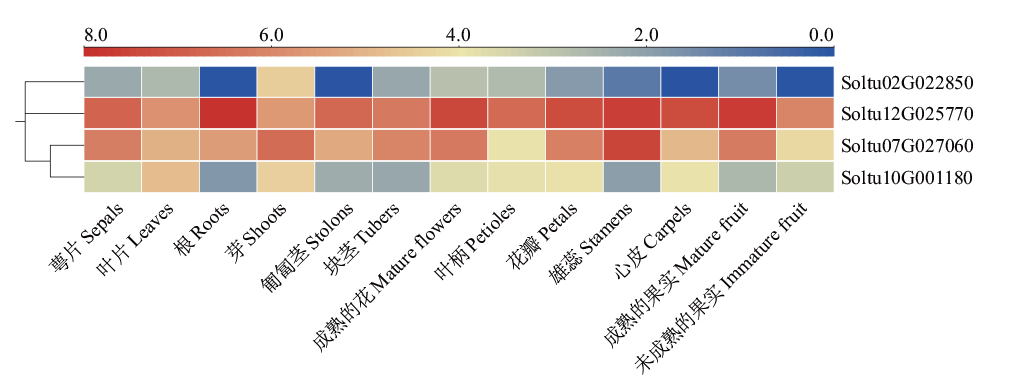
Fig. 4 StSAT gene expressions in different tissues of DM potato The expressions of four StSATs in 13 tissue sites are logarithms base 2, and heat maps are plotted using log2FPKM
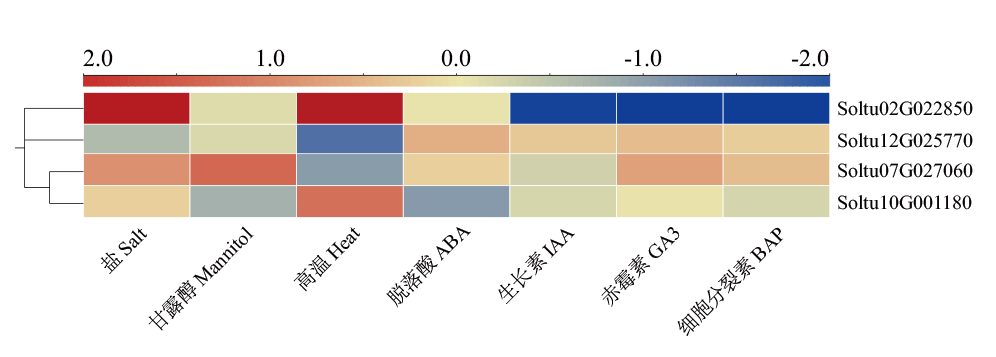
Fig. 5 StSAT gene expressions in DM potato under abiotic stress and hormone treatment The ratios of treatment to control are used for abiotic stresses(salt, mannitol, and heat stress), taken as base 2 logarithms and heat maps are plotted using log2FC
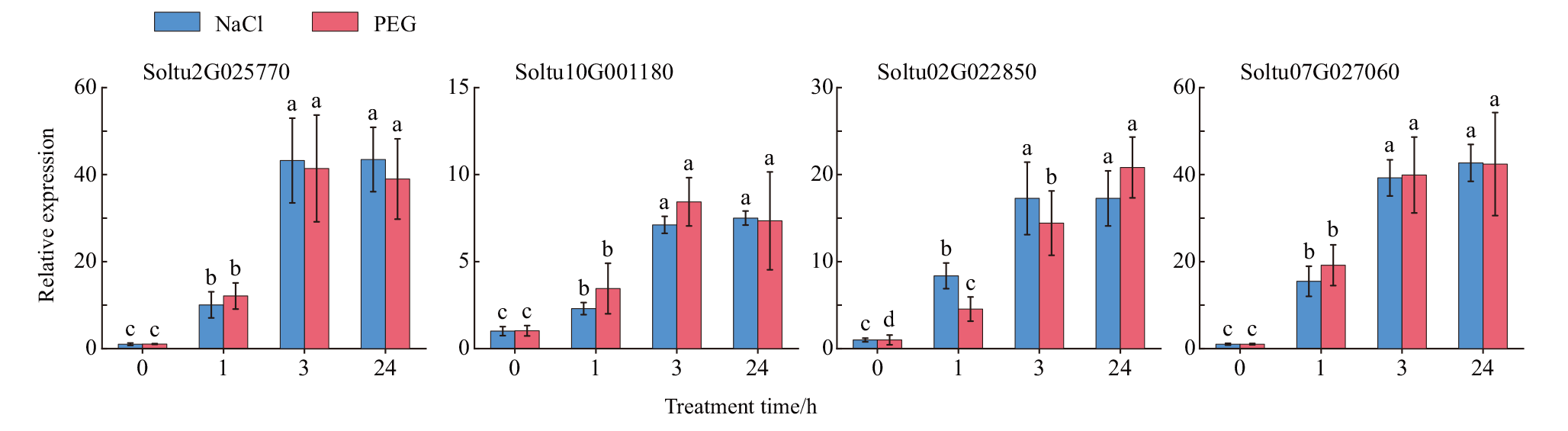
Fig. 6 Relative expressions of StSAT genes under NaCl and PEG treatment in tetraploid potato The relative expressions of four StSAT genes under NaCl and PEG treatment was analyzed by qPCR. Data are means(±SEs)from three independent biological replicates. Different letters above the bar chart indicate significant differences at P<0.05
| [1] |
Beinert H. A tribute to sulfur[J]. Eur J Biochem, 2000, 267(18): 5657-5664.
pmid: 10971575 |
| [2] | Gerber J, Lill R. Biogenesis of iron-sulfur proteins in eukaryotes: components, mechanism and pathology[J]. Mitochondrion, 2002, 2(1/2): 71-86. |
| [3] |
Saito K. Sulfur assimilatory metabolism. The long and smelling road[J]. Plant Physiol, 2004, 136(1): 2443-2450.
doi: 10.1104/pp.104.046755 pmid: 15375200 |
| [4] |
Leustek T, Saito K. Sulfate transport and assimilation in plants[J]. Plant Physiol, 1999, 120(3): 637-644.
pmid: 10398698 |
| [5] | Wirtz M, Hell R. Functional analysis of the cysteine synthase protein complex from plants: structural, biochemical and regulatory properties[J]. J Plant Physiol, 2006, 163(3): 273-286. |
| [6] |
Romero LC, Ángeles Aroca M, Laureano-Marín AM, et al. Cysteine and cysteine-related signaling pathways in Arabidopsis thaliana[J]. Mol Plant, 2014, 7(2): 264-276.
doi: 10.1093/mp/sst168 pmid: 24285094 |
| [7] | Initiative AG. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana[J]. Nature, 2000, 408(6814): 796-815. |
| [8] |
Noji M, Inoue K, Kimura N, et al. Isoform-dependent differences in feedback regulation and subcellular localization of serine acetyltransferase involved in cysteine biosynthesis from Arabidopsis thaliana[J]. J Biol Chem, 1998, 273(49): 32739-32745.
doi: 10.1074/jbc.273.49.32739 pmid: 9830017 |
| [9] |
Kawashima CG, Berkowitz O, Hell R, et al. Characterization and expression analysis of a serine acetyltransferase gene family involved in a key step of the sulfur assimilation pathway in Arabidopsis[J]. Plant Physiol, 2005, 137(1): 220-230.
pmid: 15579666 |
| [10] |
Wirtz M, Berkowitz O, Droux M, et al. The cysteine synthase complex from plants. Mitochondrial serine acetyltransferase from Arabidopsis thaliana carries a bifunctional domain for catalysis and protein-protein interaction[J]. Eur J Biochem, 2001, 268(3): 686-693.
pmid: 11168407 |
| [11] | Kumar S, Kumar N, Alam N, et al. Crystal structure of serine acetyl transferase from Brucella abortus and its complex with coenzyme A[J]. Biochim Biophys Acta, 2014, 1844(10): 1741-1748. |
| [12] |
Francois JA, Kumaran S, Jez JM. Structural basis for interaction of O-acetylserine sulfhydrylase and serine acetyltransferase in the Arabidopsis cysteine synthase complex[J]. Plant Cell, 2006, 18(12): 3647-3655.
doi: 10.1105/tpc.106.047316 pmid: 17194764 |
| [13] | Jez JM, Dey S. The cysteine regulatory complex from plants and microbes: what was old is new again[J]. Curr Opin Struct Biol, 2013, 23(2): 302-310. |
| [14] |
Watanabe M, Mochida K, Kato T, et al. Comparative genomics and reverse genetics analysis reveal indispensable functions of the serine acetyltransferase gene family in Arabidopsis[J]. Plant Cell, 2008, 20(9): 2484-2496.
doi: 10.1105/tpc.108.060335 pmid: 18776059 |
| [15] |
Xiang XL, Wu YR, Planta J, et al. Overexpression of serine acetyltransferase in maize leaves increases seed-specific methionine-rich zeins[J]. Plant Biotechnol J, 2018, 16(5): 1057-1067.
doi: 10.1111/pbi.12851 pmid: 29044890 |
| [16] | Zhou H, Chen Y, Zhai FC, et al. Hydrogen sulfide promotes rice drought tolerance via reestablishing redox homeostasis and activation of ABA biosynthesis and signaling[J]. Plant Physiol Biochem, 2020, 155: 213-220. |
| [17] | Cao MJ, Wang Z, Zhao Q, et al. Sulfate availability affects ABA levels and germination response to ABA and salt stress in Arabidopsis thaliana[J]. Plant J, 2014, 77(4): 604-615. |
| [18] |
Dominguez-Solis JR, He ZY, Lima A, et al. A cyclophilin links redox and light signals to cysteine biosynthesis and stress responses in chloroplasts[J]. Proc Natl Acad Sci USA, 2008, 105(42): 16386-16391.
doi: 10.1073/pnas.0808204105 pmid: 18845687 |
| [19] |
Ahmad N, Malagoli M, Wirtz M, et al. Drought stress in maize causes differential acclimation responses of glutathione and sulfur metabolism in leaves and roots[J]. BMC Plant Biol, 2016, 16(1): 247.
pmid: 27829370 |
| [20] | Na G, Salt DE. Differential regulation of serine acetyltransferase is involved in nickel hyperaccumulation in Thlaspi goesingense[J]. J Biol Chem, 2011, 286(47): 40423-40432. |
| [21] | Liu DM, Li M, Guo T, et al. Functional characterization of the Serine acetyltransferase family genes uncovers the diversification and conservation of cysteine biosynthesis in tomato[J]. Front Plant Sci, 2022, 13: 913856. |
| [22] | Kurt F, Filiz E, Aydın A. Genome-wide identification of serine acetyltransferase(SAT)gene family in rice(Oryza sativa)and their expressions under salt stress[J]. Mol Biol Rep, 2021, 48(9): 6277-6290. |
| [23] | Gasteiger E, Hoogland C, Gattiker A, et al. Protein identification and analysis tools on the ExPASy server[M]//The Proteomics Protocols Handbook. Totowa, NJ: Humana Press, 2005: 571-607. |
| [24] |
Hall BG. Building phylogenetic trees from molecular data with MEGA[J]. Mol Biol Evol, 2013, 30(5): 1229-1235.
doi: 10.1093/molbev/mst012 pmid: 23486614 |
| [25] |
Hu B, Jin JP, Guo AY, et al. GSDS 2.0: an upgraded gene feature visualization server[J]. Bioinformatics, 2015, 31(8): 1296-1297.
doi: 10.1093/bioinformatics/btu817 pmid: 25504850 |
| [26] | Bailey TL, Boden M, Buske FA, et al. MEME SUITE: tools for motif discovery and searching[J]. Nucleic Acids Res, 2009, 37(Web Server issue): W202-W208. |
| [27] | Chen CJ, Chen H, He YH, et al. TBtools, a Toolkit for Biologists integrating various biological data handling tools with a user-friendly interface[J]. bioRxiv, 2018. DOI: 10.1101/289660. |
| [28] | Liu Z, Liu YH, Coulter JA, et al. The WD40 gene family in potato(Solanum tuberosum L.): genome-wide analysis and identification of anthocyanin and drought-related WD40s[J]. Agronomy, 2020, 10(3): 401. |
| [29] | Li YM, Liang J, Zeng XZ, et al. Genome-wide analysis of MYB gene family in potato provides insights into tissue-specific regulation of anthocyanin biosynthesis[J]. Hortic Plant J, 2021, 7(2): 129-141. |
| [30] | Cannon SB, Mitra A, Baumgarten A, et al. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana[J]. BMC Plant Biol, 2004, 4: 10. |
| [31] | Yeon JY, Yoo SJ, Takagi H, et al. A novel mitochondrial serine O-acetyltransferase, OpSAT1, plays a critical role in sulfur metabolism in the thermotolerant methylotrophic yeast Ogataea parapolymorpha[J]. Sci Rep, 2018, 8(1): 2377. |
| [32] | Venkatesh TV, Harrigan GG, Perez T, et al. Compositional assessments of key maize populations: B73 hybrids of the nested association mapping founder lines and diverse landrace inbred lines[J]. J Agric Food Chem, 2015, 63(21): 5282-5295. |
| [33] | Genisel M, Erdal S, Kizilkaya M. The mitigating effect of cysteine on growth inhibition in salt-stressed barley seeds is related to its own reducing capacity rather than its effects on antioxidant system[J]. Plant Growth Regul, 2015, 75(1): 187-197. |
| [34] | Sadak MS, Abd El-Hameid AR, Zaki FSA, et al. Physiological and biochemical responses of soybean(Glycine max L.) to cysteine application under sea salt stress[J]. Bull Natl Res Cent, 2019, 44(1): 1. |
| [35] | Zhang R, Xu C, Bao ZL, et al. Auxin alters sodium ion accumulation and nutrient accumulation by playing protective role in salinity challenged strawberry[J]. Plant Physiol Biochem, 2021, 164: 1-9. |
| [36] | Khalid A, Aftab F. Effect of exogenous application of IAA and GA3 on growth, protein content, and antioxidant enzymes of Solanum tuberosum L. grown in vitro under salt stress[J]. Vitro Cell Dev Biol Plant, 2020, 56(3): 377-389. |
| [1] | WANG Chao, BAI Ru-qian, GUAN Jun-mei, LUO Ji-lin, HE Xue-jiao, CHI Shao-yi, MA Ling. Promotion of StHY5 in the Synthesis of SGAs during Tuber Turning-green of Potato [J]. Biotechnology Bulletin, 2024, 40(9): 113-122. |
| [2] | XIA Shi-xuan, GENG Ze-dong, ZHU Guang-tao, ZHANG Chun-zhi, LI Da-wei. Quick Detection of Potato Pollen Viability Based on Deep Learning [J]. Biotechnology Bulletin, 2024, 40(9): 123-130. |
| [3] | MAO Xiang-hong, LU Yao, FAN Xiang-bin, DU Pei-bing, BAI Xiao-dong. Genetic Diversity Analysis of Potato Varieties Based on SSR Fluorescent Marker Capillary Electrophoresis and Construction of Molecular Identity Card [J]. Biotechnology Bulletin, 2024, 40(9): 131-140. |
| [4] | YUAN Lan, HUANG Ya-nan, ZHANG Bei-ni, XIONG Yu-meng, WANG Hong-yang. High-throughput Sample Preparation Method for the Identification of Potato Ploidy Using Flow Cytometry [J]. Biotechnology Bulletin, 2024, 40(9): 141-147. |
| [5] | WU Hui-qin, WANG Yan-hong, LIU Han, SI Zheng, LIU Xue-qing, WANG Jing, YANG Yi, CHENG Yan. Identification and Expression Analysis of UGT Gene Family in Pepper [J]. Biotechnology Bulletin, 2024, 40(9): 198-211. |
| [6] | SONG Qian-na, DUAN Yong-hong, FENG Rui-yun. Establishment of CRISPR/Cas9-mediated Highly Efficient Gene Editing System in Microtubers of Potatoes [J]. Biotechnology Bulletin, 2024, 40(9): 33-41. |
| [7] | WANG Ke-ran, YAN Jun-jie, LIU Jian-feng, GAO Yu-lin. Application and Risk of RNAi Technology in Potato Insect Pest Management [J]. Biotechnology Bulletin, 2024, 40(9): 4-10. |
| [8] | ZHANG Xiao-mei, ZHOU Nan-ling, ZHANG Sai-hang, WANG Chao, SHEN Yu-long, GUAN Jun-mei, MA Ling. Cloning and Expression Analysis of StDREBs Gene in Solanum tuberosum L. [J]. Biotechnology Bulletin, 2024, 40(9): 42-50. |
| [9] | MAN Quan-cai, MENG Zi-nuo, LI Wei, CAI Xin-ru, SU Run-dong, FU Chang-qing, GAO Shun-juan, CUI Jiang-hui. Identification and Expression Analysis of AQP Gene Family in Potato [J]. Biotechnology Bulletin, 2024, 40(9): 51-63. |
| [10] | WU Juan, WU Xiao-juan, WANG Pei-jie, XIE Rui, NIE Hu-shuai, LI Nan, MA Yan-hong. Screening and Expression Analysis of ERF Gene Related to Anthocyanin Synthesis in Colored Potato [J]. Biotechnology Bulletin, 2024, 40(9): 82-91. |
| [11] | QIAO Yan, YANG Fang, REN Pan-rong, QI Wei-liang, AN Pei-pei, LI Qian, LI Dan, XIAO Jun-fei. Cloning and Function Analysis of the ScDHNS Gene of Crotonase/Enoyl-CoA Superfamily from a Wild Potato Species [J]. Biotechnology Bulletin, 2024, 40(9): 92-103. |
| [12] | SONG Bing-fang, LIU Ning, CHENG Xin-yan, XU Xiao-bin, TIAN Wen-mao, GAO Yue, BI Yang, WANG Yi. Identification of Potato G6PDH Gene Family and Its Expression Analysis in Damaged Tubers [J]. Biotechnology Bulletin, 2024, 40(9): 104-112. |
| [13] | ZHOU Ran, WANG Xing-ping, LI Yan-xia, LUORENG Zhuo-ma. Analysis of LncRNA Differential Expression in Mammary Tissue of Cows with Staphylococcus aureus Mastitis [J]. Biotechnology Bulletin, 2024, 40(8): 320-328. |
| [14] | LI Yi-jun, YANG Xiao-bei, XIA Lin, LUO Zhao-peng, XU Xin, YANG Jun, NING Qian-ji, WU Ming-zhu. Cloning and Functional Analysis of NtPRR37 Gene in Nicotiana tabacum L. [J]. Biotechnology Bulletin, 2024, 40(8): 221-231. |
| [15] | LI Yong-hui, BAO Xing-xing, DUAN Yi-ke, ZHAO Yun-xia, YU Xiang-li, CHEN Yao, ZHANG Yan-zhao. Genome-wide Identification and Expression Features Analysis of the bZIP Family in Rhododendron henanense subsp. lingbaoense [J]. Biotechnology Bulletin, 2024, 40(8): 186-198. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||