








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (10): 233-241.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0658
Previous Articles Next Articles
SONG Qian-na1,2,3( ), WU Shi-yun1, CAO Shi-jin1, DUAN Yong-hong1, FENG Rui-yun1,3(
), WU Shi-yun1, CAO Shi-jin1, DUAN Yong-hong1, FENG Rui-yun1,3( )
)
Received:2025-06-23
Online:2025-10-26
Published:2025-10-28
Contact:
FENG Rui-yun
E-mail:songqianna1007@126.com;fengruiyun1970@163.com
SONG Qian-na, WU Shi-yun, CAO Shi-jin, DUAN Yong-hong, FENG Rui-yun. Rapid Identification of Gene StHKT1’s Function via Potato Hairy Root System[J]. Biotechnology Bulletin, 2025, 41(10): 233-241.
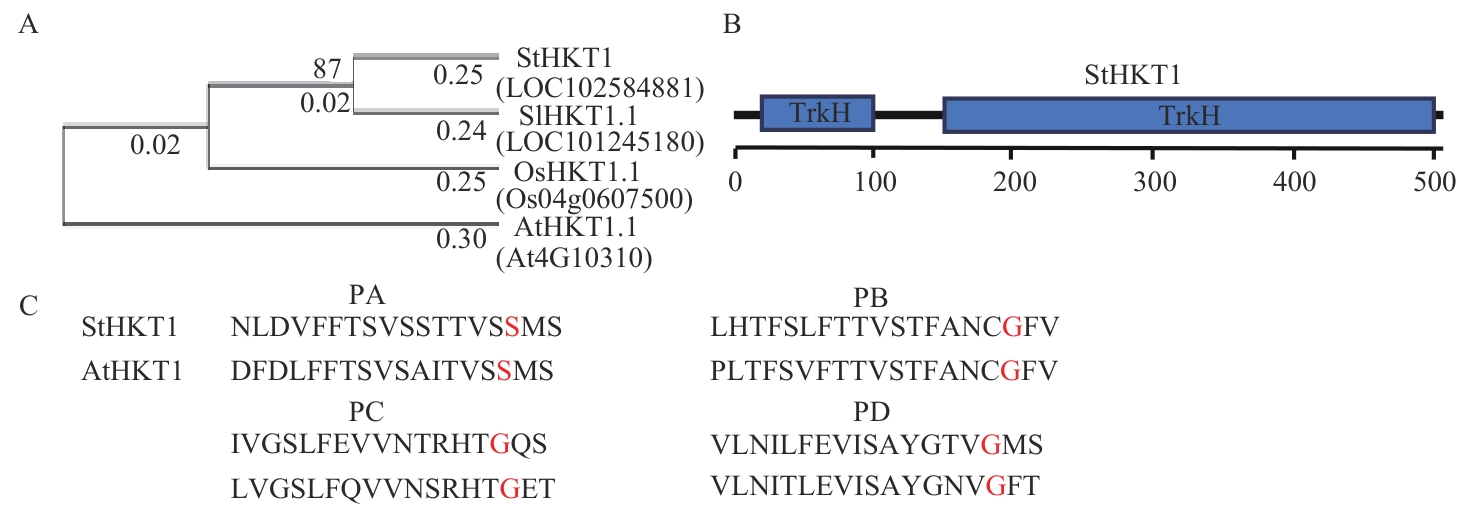
Fig. 1 Phylogenetic trees constructing and domain analysis of StHKT1 proteinA: Phylogenetic trees. B: Domain of StHKT1 protein. C: Sequence alignment of p-loop (PA, PB, PC and PD) of StHKT1 protein
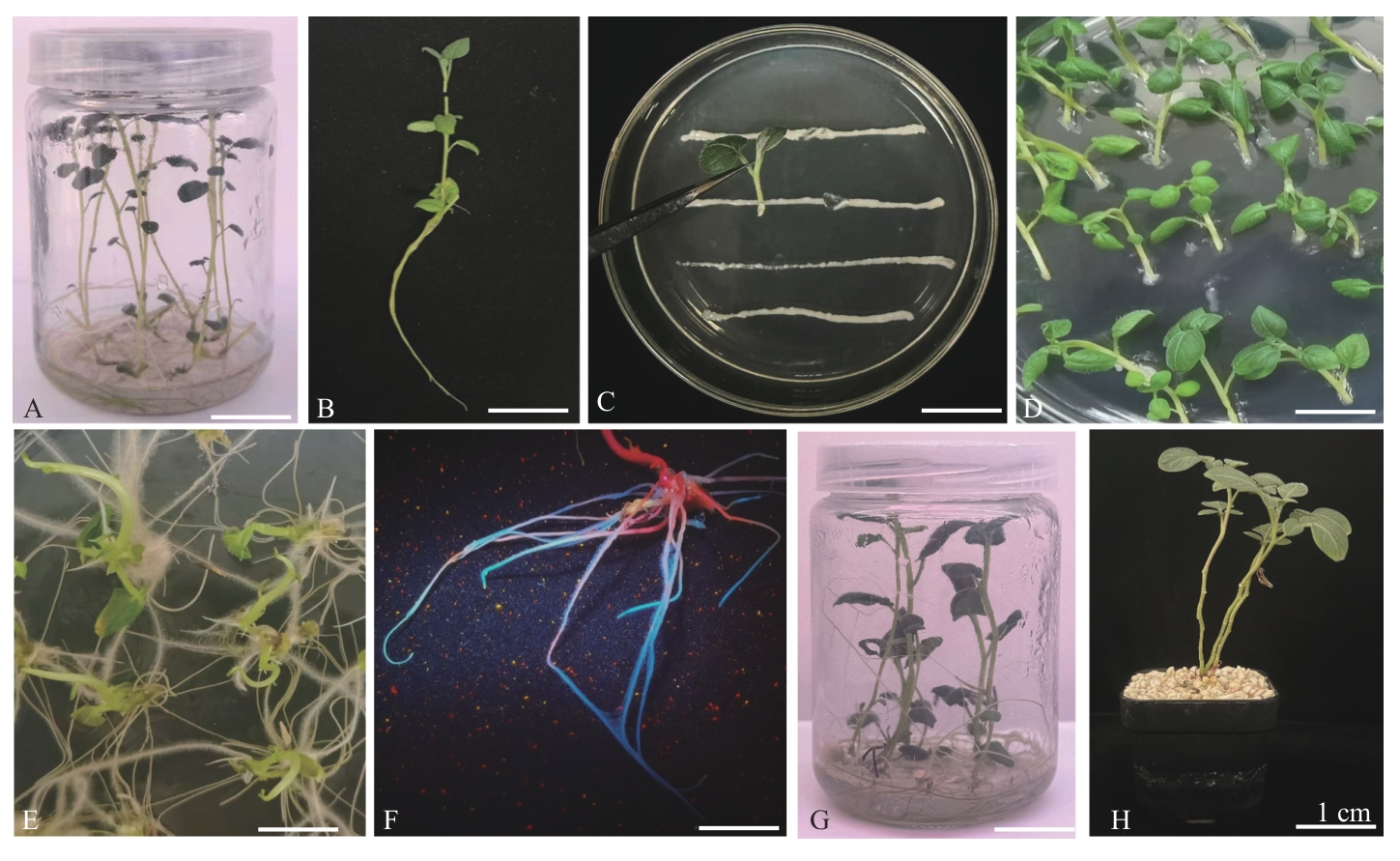
Fig. 2 Genetic transformation of potato hairy roots and obtaining of composite plantsA: Ideal potato seedings for transformation. B: Explants on terminal bud. C: Infected by MSU440 Agrobacterium rhizogenes. D: Explants co-cultured for 2 d. E: Hairy roots of induction 10 d after inoculation. F: Selection of GFP-positive hairy roots. G: Potato plants with composite. H: The composite plants cultured continuously
转化 Transformation | #1 | #2 | #3 | 毛状根诱导效率 Induction efficiency of hairy roots | 毛状根群体 Hairy root clones | #1 | #2 | #3 | 转化效率 Transformation efficiency |
|---|---|---|---|---|---|---|---|---|---|
| 侵染外植体数 | 30 | 30 | 30 | 100% | 总毛状根数 | 16 | 21 | 21 | 87.4% |
| 诱导毛状根外植体数 | 30 | 30 | 30 | 阳性毛状根数 | 13 | 18 | 20 |
Table 1 Evaluation on the induction efficiency and transformation efficiency of hairy roots
转化 Transformation | #1 | #2 | #3 | 毛状根诱导效率 Induction efficiency of hairy roots | 毛状根群体 Hairy root clones | #1 | #2 | #3 | 转化效率 Transformation efficiency |
|---|---|---|---|---|---|---|---|---|---|
| 侵染外植体数 | 30 | 30 | 30 | 100% | 总毛状根数 | 16 | 21 | 21 | 87.4% |
| 诱导毛状根外植体数 | 30 | 30 | 30 | 阳性毛状根数 | 13 | 18 | 20 |
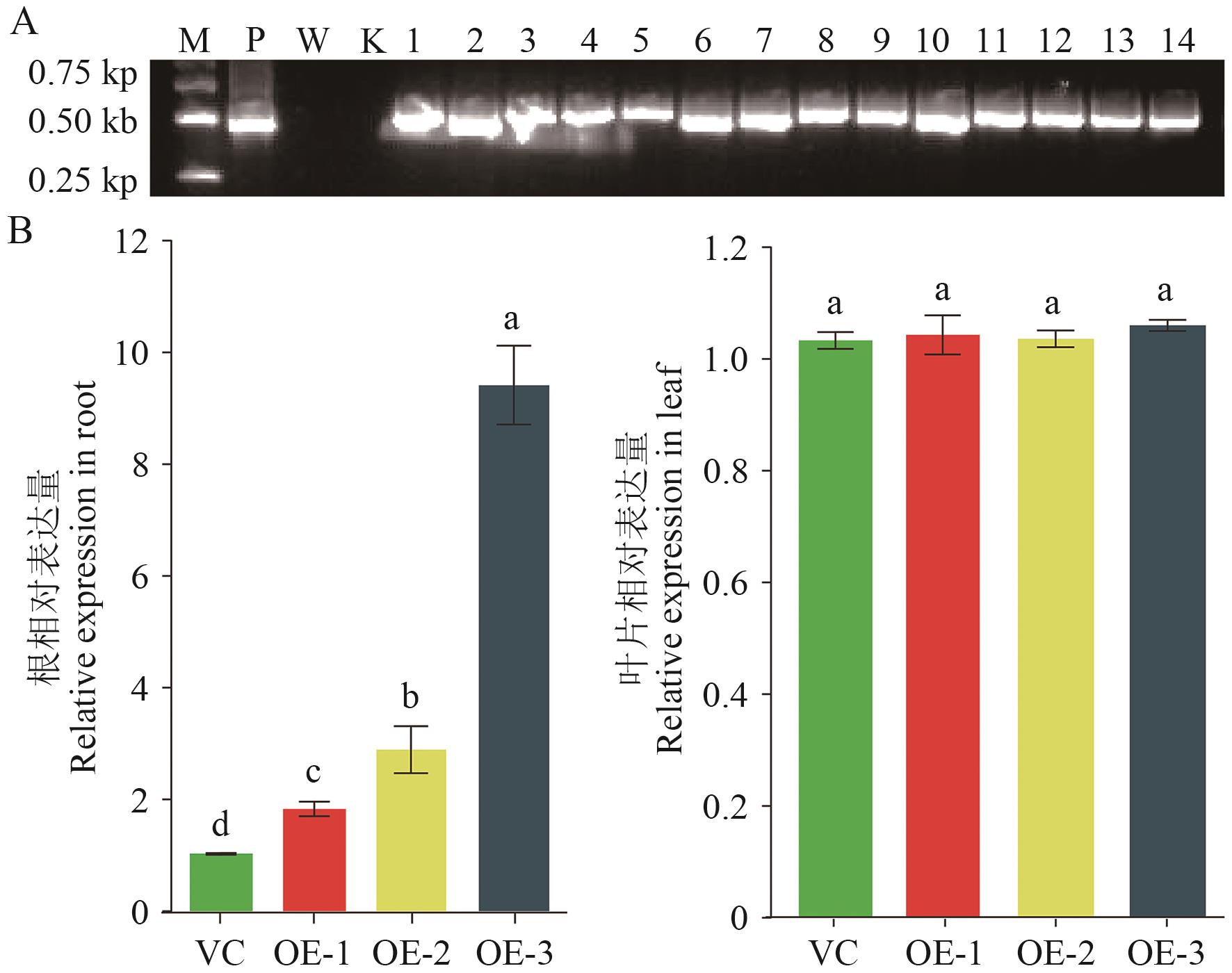
Fig. 3 Detection of relative expressions of StHKT1 in the composite potato plantsA: PCR detection of StHKT1 geneintegration. P: Plasmid; W: wild type; K: ddH2O. B: Detecting relative expression of StHKT1 by RT-qPCR. The different lowercase letters indicate P<0.05. The same below
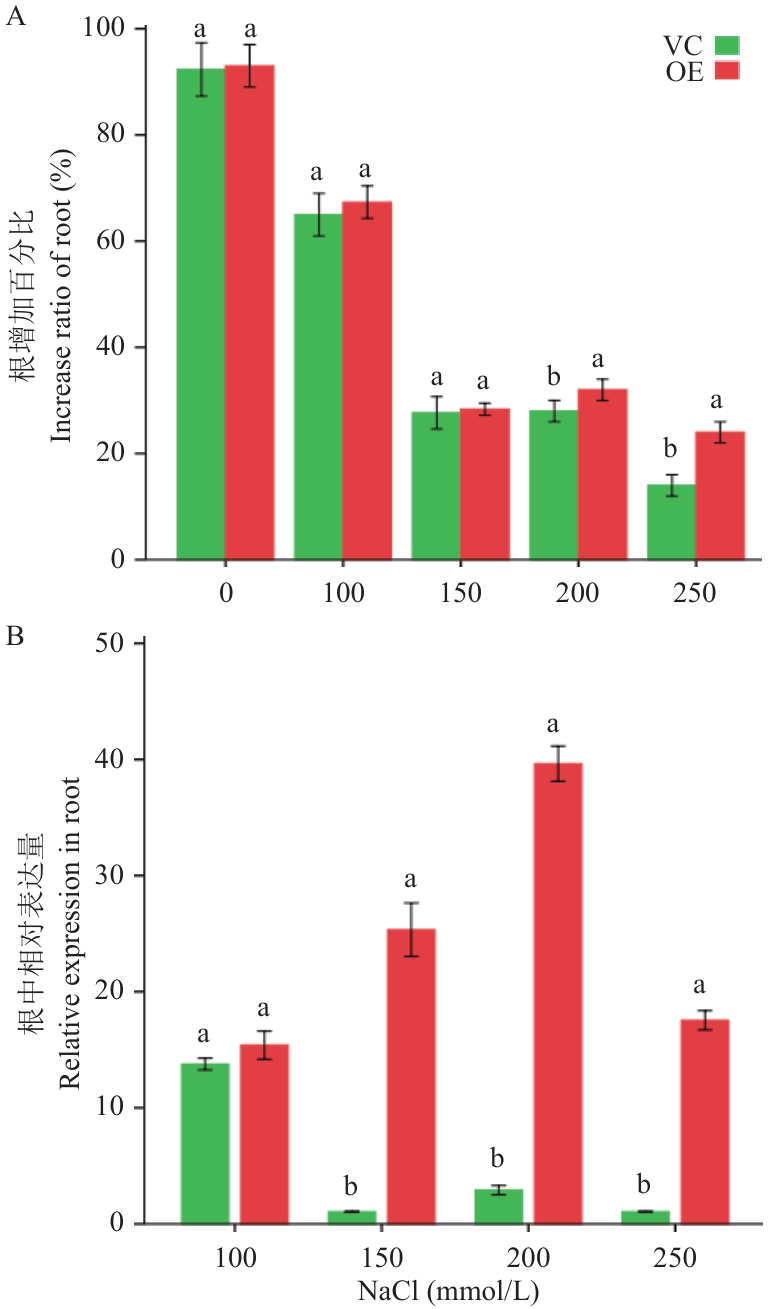
Fig. 4 Analysis of tolerance to salt and StHKT1 expression pattern of hairy roots in the composite potato plantsA: Increase ratio of hairy roots under different NaCl concentrations. B: Relative expression of StHKT1 in hairy roots under different NaCl concentrations. VC and OE contain mixed samples of hairy roots from three independent composite plants
样本 Sample | NaCl浓度 Concentrations of NaCl (mmol/L) | 根长 Length of root (cm) | 根数 No. of roots | 根鲜重 Fresh weight of root (g) | 茎长 Length of shoot (cm) | 植株鲜重 Fresh weight of plant (g) |
|---|---|---|---|---|---|---|
| VC | 0 | 9.833±0.763a | 3.000±1.000b | 0.021±0.008a | 4.567±0.551a | 0.059±0.001a |
| 100 | 5.300±0.650bc | 3.000±1.000b | 0.006±0.001b | 1.967±0.642b | 0.039±0.002bc | |
| OE | 0 | 9.600±2.970a | 5.000±0.001a | 0.025±0.021a | 5.100±0.529a | 0.080±0.012a |
| 100 | 6.900±0.360b | 4.000±1.000b | 0.007±0.002b | 2.967±0.586b | 0.059±0.008b |
Table 2 Growth indicators of the composite potato plants under salt stress
样本 Sample | NaCl浓度 Concentrations of NaCl (mmol/L) | 根长 Length of root (cm) | 根数 No. of roots | 根鲜重 Fresh weight of root (g) | 茎长 Length of shoot (cm) | 植株鲜重 Fresh weight of plant (g) |
|---|---|---|---|---|---|---|
| VC | 0 | 9.833±0.763a | 3.000±1.000b | 0.021±0.008a | 4.567±0.551a | 0.059±0.001a |
| 100 | 5.300±0.650bc | 3.000±1.000b | 0.006±0.001b | 1.967±0.642b | 0.039±0.002bc | |
| OE | 0 | 9.600±2.970a | 5.000±0.001a | 0.025±0.021a | 5.100±0.529a | 0.080±0.012a |
| 100 | 6.900±0.360b | 4.000±1.000b | 0.007±0.002b | 2.967±0.586b | 0.059±0.008b |
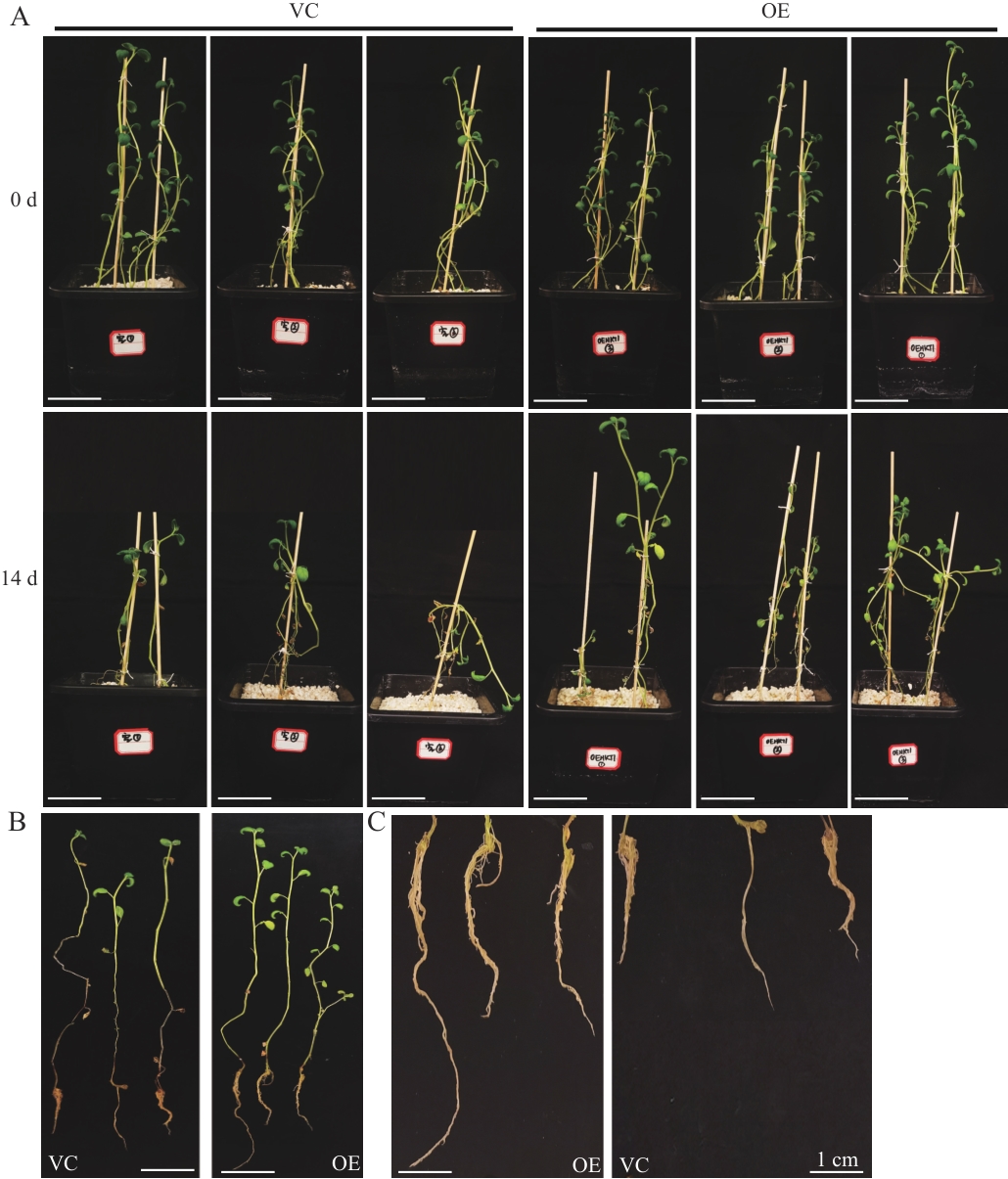
Fig. 5 Phenotypic analysis of StHKT1-overexpressed composite potato plants under salt stressA-B: Growth status of composite plants treated with salt for two weeks. C: Growth status of hairy roots of composite treated with salt for two weeks

Fig. 6 Physiological characteristics of overexpressing StHKT1 composite potato plants under salt stressA: Relative expression of StHKT1 in the hairy roots under salt stress. B: Chlorophyll content in leaf. C, E: SOD activity in leaf and hairy roots. D, F: MDA content of leaf and hairy roots. OE-1, OE-2 and OE-3 refer to three independent hairy roots from composite plants; VC and OE contain mixed samples of leaves and hairy roots from three independent composite plants
| [1] | van Zelm E, Zhang YX, Testerink C. Salt tolerance mechanisms of plants [J]. Annu Rev Plant Biol, 2020, 71: 403-433. |
| [2] | Chen ZL, Debernardi JM, Dubcovsky J, et al. Recent advances in crop transformation technologies [J]. Nat Plants, 2022, 8(12): 1343-1351. |
| [3] | Ji X, Yang B, Wang DW. Achieving plant genome editing while bypassing tissue culture [J]. Trends Plant Sci, 2020, 25(5): 427-429. |
| [4] | Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana [J]. Plant J, 1998, 16(6): 735-743. |
| [5] | Cho HJ, Farrand SK, Noel GR, et al. High-efficiency induction of soybean hairy roots and propagation of the soybean cyst nematode [J]. Planta, 2000, 210(2): 195-204. |
| [6] | Katerji N, van Hoorn JW, Hamdy A, et al. Salt tolerance classification of crops according to soil salinity and to water stress day index [J]. Agric Water Manag, 2000, 43(1): 99-109. |
| [7] | Patell RM, Prasher SO, Donnelly D, et al. Effect of initial soil salinity and subirrigation water salinity on potato tuber yield and size [J]. Agric Water Manag, 2001, 46(3): 231-239. |
| [8] | Ali A, Maggio A, Bressan RA, et al. Role and functional differences of HKT1-type transporters in plants under salt stress [J]. Int J Mol Sci, 2019, 20(5): 1059. |
| [9] | Mäser P, Hosoo Y, Goshima S, et al. Glycine residues in potassium channel-like selectivity filters determine potassium selectivity in four-loop-per-subunit HKT transporters from plants [J]. Proc Natl Acad Sci USA, 2002, 99(9): 6428-6433. |
| [10] | Henderson SW, Dunlevy JD, Wu Y, et al. Functional differences in transport properties of natural HKT1;1 variants influence shoot Na+ exclusion in grapevine rootstocks [J]. New Phytol, 2018, 217(3): 1113-1127. |
| [11] | Wang L, Liu YH, Feng SJ, et al. AtHKT1 gene regulating K+ state in whole plant improves salt tolerance in transgenic tobacco plants [J]. Sci Rep, 2018, 8: 16585. |
| [12] | Chen HT, Chen X, Gu HP, et al. GmHKT1;4, a novel soybean gene regulating Na+/K+ ratio in roots enhances salt tolerance in transgenic plants [J]. Plant Growth Regul, 2014, 73(3): 299-308. |
| [13] | Butler NM, Jansky SH, Jiang JM. First-generation genome editing in potato using hairy root transformation [J]. Plant Biotechnol J, 2020, 18(11): 2201-2209. |
| [14] | Wang L, Liu YH, Li D, et al. Improving salt tolerance in potato through overexpression of AtHKT1 gene [J]. BMC Plant Biol, 2019, 19(1): 357. |
| [15] | Ikeuchi M, Favero DS, Sakamoto Y, et al. Molecular mechanisms of plant regeneration [J]. Annu Rev Plant Biol, 2019, 70: 377-406. |
| [16] | Filipecki M, Malepszy S. Unintended consequences of plant transformation: a molecular insight [J]. J Appl Genet, 2006, 47(4): 277-286. 286. |
| [17] | Guillon S, Trémouillaux-Guiller J, Pati PK, et al. Hairy root research: recent scenario and exciting prospects [J]. Curr Opin Plant Biol, 2006, 9(3): 341-346. |
| [18] | Cao XS, Xie HT, Song ML, et al. Cut-dip-budding delivery system enables genetic modifications in plants without tissue culture [J]. Innovation, 2023, 4(1): 100345. |
| [19] | Mei GG, Chen A, Wang YR, et al. A simple and efficient in planta transformation method based on the active regeneration capacity of plants [J]. Plant Commun, 2024, 5(4): 100822. |
| [20] | Fan YL, Zhang XH, Zhong LJ, et al. One-step generation of composite soybean plants with transgenic roots by Agrobacterium rhizogenes-mediated transformation [J]. BMC Plant Biol, 2020, 20(1): 208. |
| [21] | Estrada-Navarrete G, Alvarado-Affantranger X, Olivares JE, et al. Agrobacterium rhizogenes transformation of the Phaseolus spp. a tool for functional genomics [J]. Mol Plant Microbe Interactions®, 2006, 19(12): 1385-1393. |
| [22] | Cao D, Hou WS, Liu W, et al. Overexpression of TaNHX2 enhances salt tolerance of ‘composite’ and whole transgenic soybean plants [J]. Plant Cell Tissue Organ Cult, 2011, 107(3): 541-552. |
| [23] | Ortega-Amaro MA, Rodríguez-Kessler M, Rodríguez-Hernández AA, et al. Overexpression of AtGRDP2 gene in common bean hairy roots generates vigorous plants with enhanced salt tolerance [J]. Acta Physiol Plant, 2016, 38(3): 66. |
| [24] | 柯丹霞, 霍娅娅, 刘怡, 等. 大豆TGA转录因子基因GmTGA26在盐胁迫中的功能分析 [J]. 作物学报, 2022, 48(7): 1697-1708. |
| Ke DX, Huo YY, Liu Y, et al. Functional analysis of GmTGA26 gene under salt stress in soybean [J]. Acta Agron Sin, 2022, 48(7): 1697-1708. | |
| [25] | Wang F, Chen HW, Li QT, et al. GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants [J]. Plant J, 2015, 83(2): 224-236. |
| [26] | Davenport RJ, MUÑOZ-MAYOR A, Jha D, et al. The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis [J]. Plant Cell Environ, 2007, 30(4): 497-507. |
| [27] | Sunarpi, Horie T, Motoda J, et al. Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na+ unloading from xylem vessels to xylem parenchyma cells [J]. Plant J, 2005, 44(6): 928-938. |
| [28] | Horie T, Yoshida K, Nakayama H, et al. Two types of HKT transporters with different properties of Na+ and K+ transport in Oryza sativa [J]. Plant J, 2001, 27(2): 129-138. |
| [29] | Kobayashi NI, Yamaji N, Yamamoto H, et al. OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice [J]. Plant J, 2017, 91(4): 657-670. |
| [30] | Byrt CS, Platten JD, Spielmeyer W, et al. HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1 [J]. Plant Physiol, 2007, 143(4): 1918-1928. |
| [31] | Byrt CS, Xu B, Krishnan M, et al. The Na+ transporter, TaHKT1;5-D, limits shoot Na+ accumulation in bread wheat [J]. Plant J, 2014, 80(3): 516-526. |
| [32] | Huang L, Kuang LH, Wu LY, et al. The HKT transporter HvHKT1;5 negatively regulates salt tolerance [J]. Plant Physiol, 2020, 182(1): 584-596. |
| [33] | Ali A, Raddatz N, Pardo JM, et al. HKT sodium and potassium transporters in Arabidopsis thaliana and related halophyte species [J]. Physiol Plant, 2021, 171(4): 546-558. |
| [1] | GONG Hui-ling, XING Yu-jie, MA Jun-xian, CAI Xia, FENG Zai-ping. Identification of Laccase (LAC) Gene Family in Potato (Solanum tuberosum L.) and Its Expression Analysis under Salt Stresses [J]. Biotechnology Bulletin, 2025, 41(9): 82-93. |
| [2] | LU Yao, YUAN Ping-ping, JIN Xin, MAO Xiang-hong, FAN Xiang-bin, BAI Xiao-dong. Genetic Diversity Analysis and Fingerprinting of Wild Potato and Landraces Based on SSR Markers [J]. Biotechnology Bulletin, 2025, 41(9): 94-104. |
| [3] | LI Xia, ZHANG Ze-wei, LIU Ze-jun, WANG Nan, GUO Jiang-bo, XIN Cui-hua, ZHANG Tong, JIAN Lei. Cloning and Functional Study of Transcription Factor StMYB96 in Potato [J]. Biotechnology Bulletin, 2025, 41(7): 181-192. |
| [4] | WANG Fang, QIAO Shuai, SONG Wei, CUI Peng-juan, LIAO An-zhong, TAN Wen-fang, YANG Song-tao. Genome-wide Identification of the IbNRT2 Gene Family and Its Expression in Sweet Potato [J]. Biotechnology Bulletin, 2025, 41(7): 193-204. |
| [5] | LUO Ji-lin, LI Jin-ye, JIA Yu-xin. Identification and Functional Analysis of Gravity Response Regulatory Genes in Potato [J]. Biotechnology Bulletin, 2025, 41(6): 109-118. |
| [6] | XU Hui-zhen, SHANTWANA Ghimire, RAJU Kharel, YUE Yun, SI Huai-jun, TANG Xun. Analysis of the Potato SUMO E3 Ligase Gene Family and Cloning and Expression Pattern of StSIZ1 [J]. Biotechnology Bulletin, 2025, 41(6): 119-129. |
| [7] | DUAN Yong-hong, YANG Xin, YU Guan-qun, XIA Jun-jun, SONG Lu-shuai, BAI Xiao-dong, PENG Suo-tang. Genetic Diversity and Principal Component Analysis of 125 Potato Germplasm Resources [J]. Biotechnology Bulletin, 2025, 41(6): 130-143. |
| [8] | SONG Hui-yang, SU Bao-jie, LI Jing-hao, MEI Chao, SONG Qian-na, CUI Fu-zhu, FENG Rui-yun. Cloning and Functional Analysis of the StAS2-15 Gene in Potato under Salt Stress [J]. Biotechnology Bulletin, 2025, 41(5): 119-128. |
| [9] | WEN Bo-lin, WAN Min, HU Jian-jun, WANG Ke-xiu, JING Sheng-lin, WANG Xin-yue, ZHU Bo, TANG Ming-xia, LI Bing, HE Wei, ZENG Zi-xian. Establishment of Genetic Transformation and Gene Editing System for a Potato Cultivar Chuanyu 50 [J]. Biotechnology Bulletin, 2025, 41(4): 88-97. |
| [10] | LIU Tao, WANG Zhi-qi, WU Wen-bo, SHI Wen-ting, WANG Chao-nan, DU Chong, YANG Zhong-min. Identification and Expression Analysis of the GRAM Gene Family in Potato [J]. Biotechnology Bulletin, 2025, 41(4): 145-155. |
| [11] | ZHANG Yi-xuan, MA Yu, WANG Tong-tong, SHENG Su-ao, SONG Jia-feng, LYU Zhao-yan, ZHU Xiao-biao, HOU Hua-lan. Genome-wide Identification and Expression Profiles of DIR Gene Family in Potato [J]. Biotechnology Bulletin, 2025, 41(3): 123-136. |
| [12] | YU Ting, HUANG Dan-dan, ZHU Yan-hui, YANG Mei-hong, AI Ju, GAO Dong-li. Screening and Interaction Verification of Transcription Factors Stpatatin 05 Gene in Potato [J]. Biotechnology Bulletin, 2025, 41(3): 137-145. |
| [13] | QIN Yue, YANG Yan, ZHANG Lei, LU Li-li, LI Xian-ping, JIANG Wei. Identification and Comparative Analysis of the StGAox Genes in Diploid and Tetraploid Potatoes [J]. Biotechnology Bulletin, 2025, 41(3): 146-160. |
| [14] | LYU Ji-min, LIU Wei, SUN Min, LI Hong-shun, PENG Zhen-xing, QIU Peng-fei, ZHU Qi-li. Screening, Identification and Optimization of Fermentation Conditions of Antagonistic Bacteria against Potato Early Blight [J]. Biotechnology Bulletin, 2025, 41(10): 175-185. |
| [15] | SHEN Peng, GAO Ya-Bin, DING Hong. Identification and Expression Analysis of SAT Gene Family in Potato(Solanum tuberosum L.) [J]. Biotechnology Bulletin, 2024, 40(9): 64-73. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||