








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (2): 139-149.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0584
LIU Fang1,2( ), DU Qian-qian1, HE Hao1, XIAO Gang3, YAN Zhong-yuan1(
), DU Qian-qian1, HE Hao1, XIAO Gang3, YAN Zhong-yuan1( ), HAO Xiao-hua1(
), HAO Xiao-hua1( )
)
Received:2024-06-17
Online:2025-02-26
Published:2025-02-28
Contact:
YAN Zhong-yuan, HAO Xiao-hua
E-mail:g5n2a5f@163.com;7637188893@qq.com;14771417@qq.com
LIU Fang, DU Qian-qian, HE Hao, XIAO Gang, YAN Zhong-yuan, HAO Xiao-hua. Mechanism of miR172b/c-BnMSH7.A1 Module Responding to Cu 2+ Stress in Brassica napus[J]. Biotechnology Bulletin, 2025, 41(2): 139-149.
引物名称 Primer name | 正向引物 Forward primer (5′-3′) | 反向引物 Reward primer (5′-3′) | 用途 Purpose |
|---|---|---|---|
| DL-MSH7A1 | CGTCGCAAGCTACTCAGCGT | CACATTTCGCTCCACAGAG C | RT-qPCR检测BnMSH7.A1表达量 Detect the expression level of BnMSH7.A1 by RT-qPCR |
| DL-MSH7C1 | GAAACCGTCGCAAGCTTCAG | CATTTCGCTCCACAGAG | RT-qPCR检测BnMSH7.C1表达量 Detect the expression level of BnMSH7.C1 by RT-qPCR |
| DLpre-miR172b | TGGCTTTCTGAATCCTCTTCC | GATGCTGCATCTGCAACTACC | RT-qPCR检测pre-miR172b表达量 Detect the expression level of pre-miR172b by RT-qPCR |
| DLpre-miR172c | GCCGGTAGTTGCAGATGC | GCTGATGCAGCATCATCAAG | RT-qPCR检测miR172c前体表达量 Detect the expression level of pre-miR172c by RT-qPCR |
| Actin | CTGGTGATGGTGTGTCTCACAC | GTTGTCTCATGGATTCCAGGAG | 内参基因 Reference gene |
| DLmiR172b/c | GGAATCTTGATGATGCTGCAT | 随机引物Random primer | RT-qPCR检测miR172b/c表达量 Detect the expression level of miR172b/c by RT-qPCR |
| U6 | CGATAAAATTGGAACGATACAGA | ATTTGGACCATTTCTCGATTTGT | 内参基因 Reference gene |
| G172b | CCG | G | 克隆pre-miR172b序列并构建过表达载体 Clone the pre-miR172b sequence and construct over-expression vector |
| G172c | CCG | G | 克隆pre-miR172c序列并构建过表达载体 Clone the pre-miR172c sequence and construct over-expression vector |
Table 1 Primers used in this article
引物名称 Primer name | 正向引物 Forward primer (5′-3′) | 反向引物 Reward primer (5′-3′) | 用途 Purpose |
|---|---|---|---|
| DL-MSH7A1 | CGTCGCAAGCTACTCAGCGT | CACATTTCGCTCCACAGAG C | RT-qPCR检测BnMSH7.A1表达量 Detect the expression level of BnMSH7.A1 by RT-qPCR |
| DL-MSH7C1 | GAAACCGTCGCAAGCTTCAG | CATTTCGCTCCACAGAG | RT-qPCR检测BnMSH7.C1表达量 Detect the expression level of BnMSH7.C1 by RT-qPCR |
| DLpre-miR172b | TGGCTTTCTGAATCCTCTTCC | GATGCTGCATCTGCAACTACC | RT-qPCR检测pre-miR172b表达量 Detect the expression level of pre-miR172b by RT-qPCR |
| DLpre-miR172c | GCCGGTAGTTGCAGATGC | GCTGATGCAGCATCATCAAG | RT-qPCR检测miR172c前体表达量 Detect the expression level of pre-miR172c by RT-qPCR |
| Actin | CTGGTGATGGTGTGTCTCACAC | GTTGTCTCATGGATTCCAGGAG | 内参基因 Reference gene |
| DLmiR172b/c | GGAATCTTGATGATGCTGCAT | 随机引物Random primer | RT-qPCR检测miR172b/c表达量 Detect the expression level of miR172b/c by RT-qPCR |
| U6 | CGATAAAATTGGAACGATACAGA | ATTTGGACCATTTCTCGATTTGT | 内参基因 Reference gene |
| G172b | CCG | G | 克隆pre-miR172b序列并构建过表达载体 Clone the pre-miR172b sequence and construct over-expression vector |
| G172c | CCG | G | 克隆pre-miR172c序列并构建过表达载体 Clone the pre-miR172c sequence and construct over-expression vector |
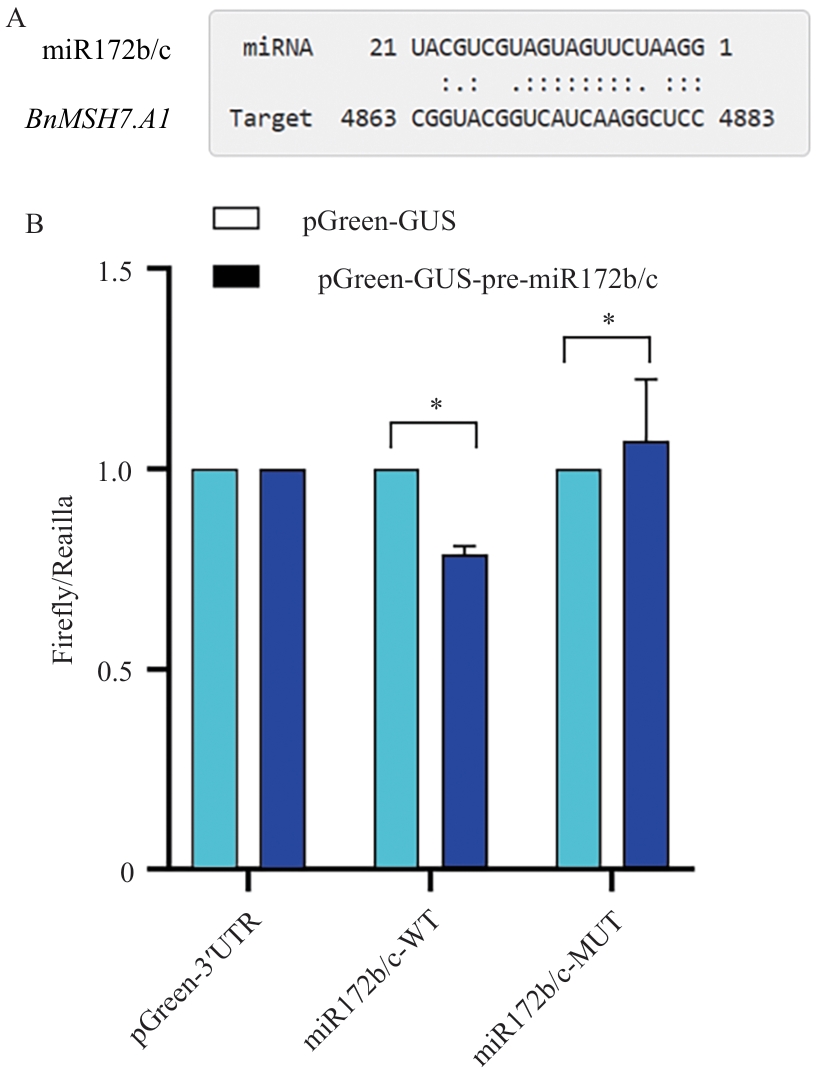
Fig. 3 Pairing of BnMSH7.A1 with miR172b/c and double luciferase validation resultsA: The predicted pairing sites between BnMSH7.A1 and miR172b/c. B: Validation of the interaction between miR172b/c and BnMSH7.A1 using dual-luciferase assay. *P<0.05, **P<0.01. The same below

Fig. 4 Expressions of BnMSH7.A1 and miR172b/c after over-expressionA: Expression variation of BnMSH7.A1 in rape cotyledons after the over expression of pre-miR172b or pre-miR172c. B: Expression variation of miR172b and miR172c in rape cotyledons after the over expression of pre-miR172b or pre-miR172c
相关性 Correlation | 根 Root | 叶 Leaf | |||
|---|---|---|---|---|---|
| r | P | r | P | ||
| Pre-miR172b:miR172b | 0.61 | 0.02 | -0.45 | 0.02 | |
| Pre-miR172c:miR172c | 0.73 | 0.03 | -0.31 | 0.04 | |
| miR172b/c:BnMSH7.A1 | 0.52 | 0 | -0.78 | 0.04 | |
Table 2 Correlation analysis
相关性 Correlation | 根 Root | 叶 Leaf | |||
|---|---|---|---|---|---|
| r | P | r | P | ||
| Pre-miR172b:miR172b | 0.61 | 0.02 | -0.45 | 0.02 | |
| Pre-miR172c:miR172c | 0.73 | 0.03 | -0.31 | 0.04 | |
| miR172b/c:BnMSH7.A1 | 0.52 | 0 | -0.78 | 0.04 | |
| 1 | Filipič M. Mechanisms of cadmium induced genomic instability [J]. Mutat Res, 2012, 733(1/2): 69-77. |
| 2 | Wang HT, He L, Song J, et al. Cadmium-induced genomic instability in Arabidopsis: molecular toxicological biomarkers for early diagnosis of cadmium stress [J]. Chemosphere, 2016, 150: 258-265. |
| 3 | Cui WN, Wang HT, Song J, et al. Cell cycle arrest mediated by Cd-induced DNA damage in Arabidopsis root tips [J]. Ecotoxicol Environ Saf, 2017, 145: 569-574. |
| 4 | Marti TM, Kunz C, Fleck O. DNA mismatch repair and mutation avoidance pathways [J]. J Cell Physiol, 2002, 191(1): 28-41. |
| 5 | Golubov A, Yao YL, Maheshwari P, et al. Microsatellite instability in Arabidopsis increases with plant development [J]. Plant Physiol, 2010, 154(3): 1415-1427. |
| 6 | Dzantiev L, Constantin N, Genschel J, et al. A defined human system that supports bidirectional mismatch-provoked excision [J]. Mol Cell, 2004, 15(1): 31-41. |
| 7 | Gomez RL, Galles C, Spampinato CP. High-level production of MSH2 from Arabidopsis thaliana: a DNA mismatch repair system key subunit [J]. Mol Biotechnol, 2011, 47(2): 120-129. |
| 8 | Culligan KM, Hays JB. Arabidopsis MutS homologs-AtMSH2, AtMSH3, AtMSH6, and a novel AtMSH7-form three distinct protein heterodimers with different specificities for mismatched DNA [J]. Plant Cell, 2000, 12(6): 991-1002. |
| 9 | Guo ML, Gao WX, Yu XJ, et al. Data mining of Arabidopsis thaliana salt-response proteins based on bioinformatics analysis [J]. Plant OMICS, 2012, 5(2): 75-78. |
| 10 | Chirinos-Arias MC, Spampinato CP. Role of the mismatch repair protein MSH7 in Arabidopsis adaptation to acute salt stress [J]. Plant Physiol Biochem, 2021, 169: 280-290. |
| 11 | 孙晓霞, 宋有涛, 李照令, 等. Cu胁迫对拟南芥幼苗错配修复基因表达的影响 [J]. 生态学杂志, 2013, 32(8): 1973-1979. |
| Sun XX, Song YT, Li ZL, et al. Effects of copper stress on the expression of DNA mismatch repair genes in Arabidopsis thaliana plantlets [J]. Chin J Ecol, 2013, 32(8): 1973-1979. | |
| 12 | 张幸媛, 田宇豪, 秦玉芝, 等. miR169在植物生长发育与非生物胁迫响应中的作用 [J]. 植物遗传资源学报, 2021, 22(4): 900-909. |
| Zhang XY, Tian YH, Qin YZ, et al. The role of miR169 family members in the processes of growth, development and abiotic stress response in planta [J]. J Plant Genet Resour, 2021, 22(4): 900-909. | |
| 13 | Orangi E, Motovali-Bashi M. Evaluation of miRNA-9 and miRNA-34a as potential biomarkers for diagnosis of breast cancer in Iranian women [J]. Gene, 2019, 687: 272-279. |
| 14 | Ye L, Jiang T, Shao HZ, et al. MiR-1290 is a biomarker in DNA-mismatch-repair-deficient colon cancer and promotes resistance to 5-fluorouracil by directly targeting hMSH2 [J]. Mol Ther Nucleic Acids, 2017, 7: 453-464. |
| 15 | Zhou ZS, Song JB, Yang ZM. Genome-wide identification of Brassica napus microRNAs and their targets in response to cadmium [J]. J Exp Bot, 2012, 63(12): 4597-4613. |
| 16 | Budak H, Kantar M, Bulut R, et al. Stress responsive miRNAs and isomiRs in cereals [J]. Plant Sci, 2015, 235: 1-13. |
| 17 | Ding YF, Zhu C. The role of microRNAs in copper and cadmium homeostasis [J]. Biochem Biophys Res Commun, 2009, 386(1): 6-10. |
| 18 | Ding YF, Chen Z, Zhu C. Microarray-based analysis of cadmium-responsive microRNAs in rice (Oryza sativa) [J]. J Exp Bot, 2011, 62(10): 3563-3573. |
| 19 | Qiu ZB, Hai BZ, Guo JL, et al. Characterization of wheat miRNAs and their target genes responsive to cadmium stress [J]. Plant Physiol Biochem, 2016, 101: 60-67. |
| 20 | 成智博, 王鹤潼, 赵强, 等. 拟南芥miRNA172b-5p、miRNA172e-5p和miRNA472-3p靶向MSH6基因参与Cd应激响应 [J]. 生态学杂志, 2019, 38(12): 3738-3746. |
| Cheng ZB, Wang HT, Zhao Q, et al. MiRNA172b-5p, miRNA172e-5p and miRNA472-3p responded to Cd stress by targeting MSH6 gene in Arabidopsis thaliana [J]. Chin J Ecol, 2019, 38(12): 3738-3746. | |
| 21 | Liu QK, Axtell MJ. Quantitating plant microRNA-mediated target repression using a dual-luciferase transient expression system [J]. Methods Mol Biol, 2015, 1284: 287-303. |
| 22 | 谭小力, 诸葛锐军, 李冠英, 等. 农杆菌介导的油菜子叶瞬时表达 [J]. 生物学杂志, 2012, 29(6): 93-96. |
| Tan XL, Zhuge RJ, Li GY, et al. An agrobacterium-mediated transit transformation system in Brassica napus cotyledon [J]. J Microbiol, 2012, 29(6): 93-96. | |
| 23 | 宋玉芳, 许华夏, 任丽萍, 等. 重金属对西红柿种子发芽与根伸长的抑制效应 [J]. 中国环境科学, 2001, 21(5): 390-394. |
| Song YF, Xu HX, Ren LP, et al. Inhibition effect of heavy metals in soil on the inhibition of seed germination and root elongation of tomatoes [J]. China Environ Sci, 2001, 21(5): 390-394. | |
| 24 | Chirinos-Arias MC, Spampinato CP. Growth and development of AtMSH7 mutants in Arabidopsis thaliana [J]. Plant Physiol Biochem, 2020, 146: 329-336. |
| 25 | Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology [J]. Annu Rev Biochem, 1996, 65: 101-133. |
| 26 | Umar A, Buermeyer AB, Simon JA, et al. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis [J]. Cell, 1996, 87(1): 65-73. |
| 27 | Wu S, Culligan K, Lamers M, et al. Dissimilar mispair-recognition spectra of Arabidopsis DNA-mismatch-repair proteins MSH2·MSH6 (MutSα) and MSH2·MSH7 (MutSγ) [J]. Nucleic Acids Res, 2003, 31(20): 6027-6034. |
| 28 | Sugawara N, Pâques F, Colaiácovo M, et al. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination [J]. Proc Natl Acad Sci USA, 1997, 94(17): 9214-9219. |
| 29 | Horwath M, Kramer W, Kunze R. Structure and expression of the Zea mays mutS-homologs Mus1 and Mus2 [J]. Theor Appl Genet, 2002, 105(2/3): 423-430. |
| 30 | Tam SM, Samipak S, Britt A, et al. Characterization and comparative sequence analysis of the DNA mismatch repair MSH2 and MSH7 genes from tomato [J]. Genetica, 2009, 137(3): 341-354. |
| 31 | Cao X, Wang HT, Zhuang DF, et al. Roles of MSH2 and MSH6 in cadmium-induced G2/M checkpoint arrest in Arabidopsis roots [J]. Chemosphere, 2018, 201: 586-594. |
| 32 | 张柳伟. miR395调节油菜(Brassica napus)耐镉功能的研究 [D]. 南京: 南京农业大学, 2012. |
| Zhang LW. Study on the regulation of miR395 on cadmium tolerance of Brassica napus [D]. Nanjing: Nanjing Agricultural University, 2012. | |
| 33 | 肖莉, 刘春, 向世鹏, 等. microRNAs在植物响应金属毒性中的作用 [J]. 衡阳师范学院学报, 2013, 34(3): 113-117. |
| Xiao L, Liu C, Xiang SP, et al. The role of microRNAs in plant response to metal toxicity [J]. J Hengyang Norm Univ, 2013, 34(3): 113-117. |
| [1] | YANG Zhi-xiao, HOU Qian, LIU Guo-quan, LU Zhi-gang, CAO Yi, GOU Jian-yu, WANG Yi, LIN Ying-chao. Responses of Rubisco and Rubisco Activase in Different Resistant Tobacco Strains to Brown Spot Stress [J]. Biotechnology Bulletin, 2023, 39(9): 202-212. |
| [2] | LI Bo, LIU He-xia, CHEN Yu-ling, ZHOU Xing-wen, ZHU Yu-lin. Cloning, Subcellular Localization and Expression Analysis of CnbHLH79 Transcription Factor from Camellia nitidissima [J]. Biotechnology Bulletin, 2023, 39(8): 241-250. |
| [3] | REN Li, QIAO Shu-ting, GE Chen-hui, WEI Zi-tong, XU Chen-xi. Identification and Expression Analysis of Spinach PSY Gene Family [J]. Biotechnology Bulletin, 2023, 39(12): 169-178. |
| [4] | YU Guo-hong, LIU Peng-cheng, LI Lei, LI Ming-zhe, CUI Hai-ying, HAO Hong-bo, GUO An-qiang. Physiological Responses of Potato in Different Genotypes to Drought Stress [J]. Biotechnology Bulletin, 2022, 38(5): 56-63. |
| [5] | LI Ai-guo, LI Ji-ming, LI He-ping, LIU Gui-hua, SONG Cong-min, WU Jun-yan. Selection of Winter Rape Varieties Suitable for Cold and Dry Areas in Hebei Province [J]. Biotechnology Bulletin, 2020, 36(1): 95-100. |
| [6] | WANG Xue-han, MA Qiang, TIAN Yuan, HU Jing, LIU Hui-rong. Cultivable Myxobacteria and Their Antibiotic Activities in the Hulun Buir Area of Inner Mongolia [J]. Biotechnology Bulletin, 2019, 35(9): 224-233. |
| [7] | LI Biao, ZHANG Rui-ying, WANG Xiao-qi, ZHANG Cun-fang, DUAN Zi-yuan. Microsatellite Polymorphism and Its Correlation Analysis with Body Size Traits of Tan Sheep [J]. Biotechnology Bulletin, 2019, 35(6): 131-137. |
| [8] | HUANG Long, WU Ben-li, HE Ji-xiang, CHEN Jing, SONG Guang-tong, WANG Xiang, ZHANG Ye, WU Song. SNP Identification of MyoD1 Gene and Its Correlation with Growth Traits in Pelodiscus sinensis [J]. Biotechnology Bulletin, 2019, 35(4): 76-81. |
| [9] | WANG Yan-xin, LIAO Yuan-yuan, A Yimuguli, QI Ao-qiong, LI Hai-jian, XU Hong-wei, YANG Ju-tian, CAI Yong. Correlation Analysis Between LHX3 Gene Polymorphism and Growth Traits in Three Sheep Breeds [J]. Biotechnology Bulletin, 2019, 35(10): 162-168. |
| [10] | JIANG Ke-ren, MA Zheng, ZHENG Hang, LIU Xiao-Jun. Review on the Application of Integrated Transcriptome and Proteome Analysis in Biology [J]. Biotechnology Bulletin, 2018, 34(12): 50-55. |
| [11] | ZHONG Shu-xia DENG Jie WEI Chun-hui LUO Hui-bo WAN Shi-lü HUANG Zhi-guo. Correlations Between Microbial Community and Physicochemical Indexes in Pit Mud of Different Ages [J]. Biotechnology Bulletin, 2016, 32(7): 119-125. |
| [12] | Niu Junqi, Huang Jingli, Zhao Wenhui, Yang Litao, Li Yangrui. Correlations Between Sucrose Accumulation and Activities of SPS, SS at Processing Maturing Stage of Sugarcane [J]. Biotechnology Bulletin, 2015, 31(9): 105-110. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||