








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (3): 190-201.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0820
SONG Shu-yi1,2( ), JIANG Kai-xiu2, LIU Huan-yan2, HUANG Ya-cheng2(
), JIANG Kai-xiu2, LIU Huan-yan2, HUANG Ya-cheng2( ), LIU Lin-ya2(
), LIU Lin-ya2( )
)
Received:2024-08-23
Online:2025-03-26
Published:2025-03-20
Contact:
HUANG Ya-cheng, LIU Lin-ya
E-mail:ishuyisong@163.com;yachenghuang1314@126.com;liulinya913@126.com
SONG Shu-yi, JIANG Kai-xiu, LIU Huan-yan, HUANG Ya-cheng, LIU Lin-ya. Identification of the TCP Gene Family in Actinidia chinensis var. Hongyang and Their Expression Analysis in Fruit[J]. Biotechnology Bulletin, 2025, 41(3): 190-201.

Fig. 1 Phylogenetic analysis (A) of TCP transcription factors in A. chinensis (Ac), A. thaliana (At), F. vesca (Fv), Ananas comosus (Aco) and P. Granatum L (Pg) and amino acid sequence comparison of conserved structural domains of AcTCPs (B)
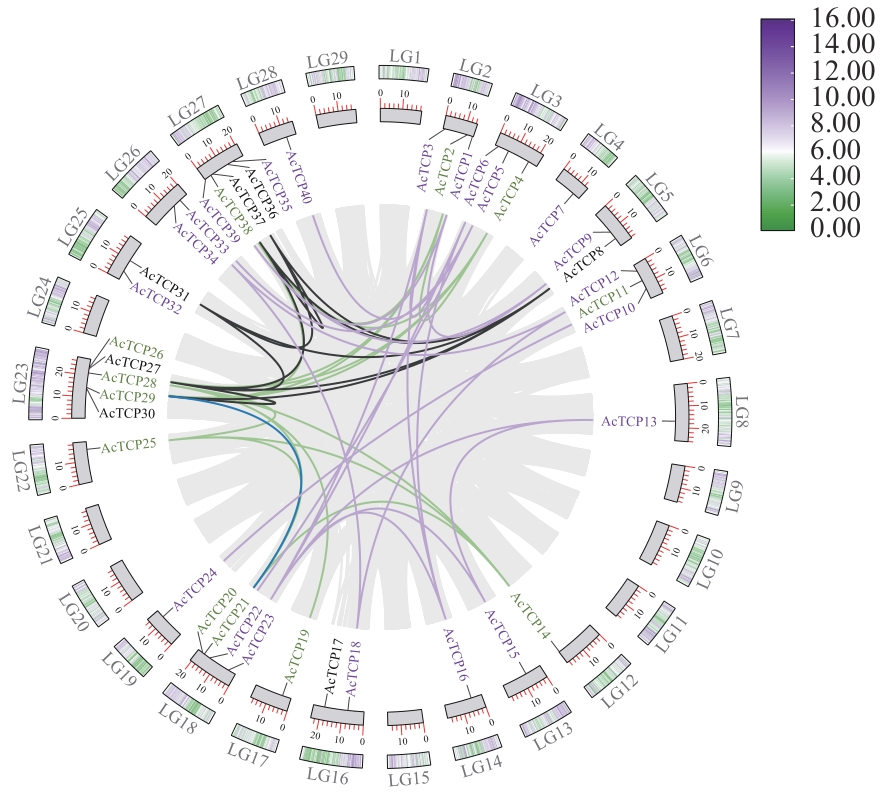
Fig. 4 Covariance analysis of AcTCPs genesPurple, green and black gene names indicate PFC genes, CIN genes and CYC/TB1 genes, respectively. Purple, green and black lines indicate intrachromosomal covariance of PFC genes, CIN genes and CYC/TB1 genes, respectively. The blue line indicates the intrachromosomal covariance between the CIN gene and the CYC/TB1 gene
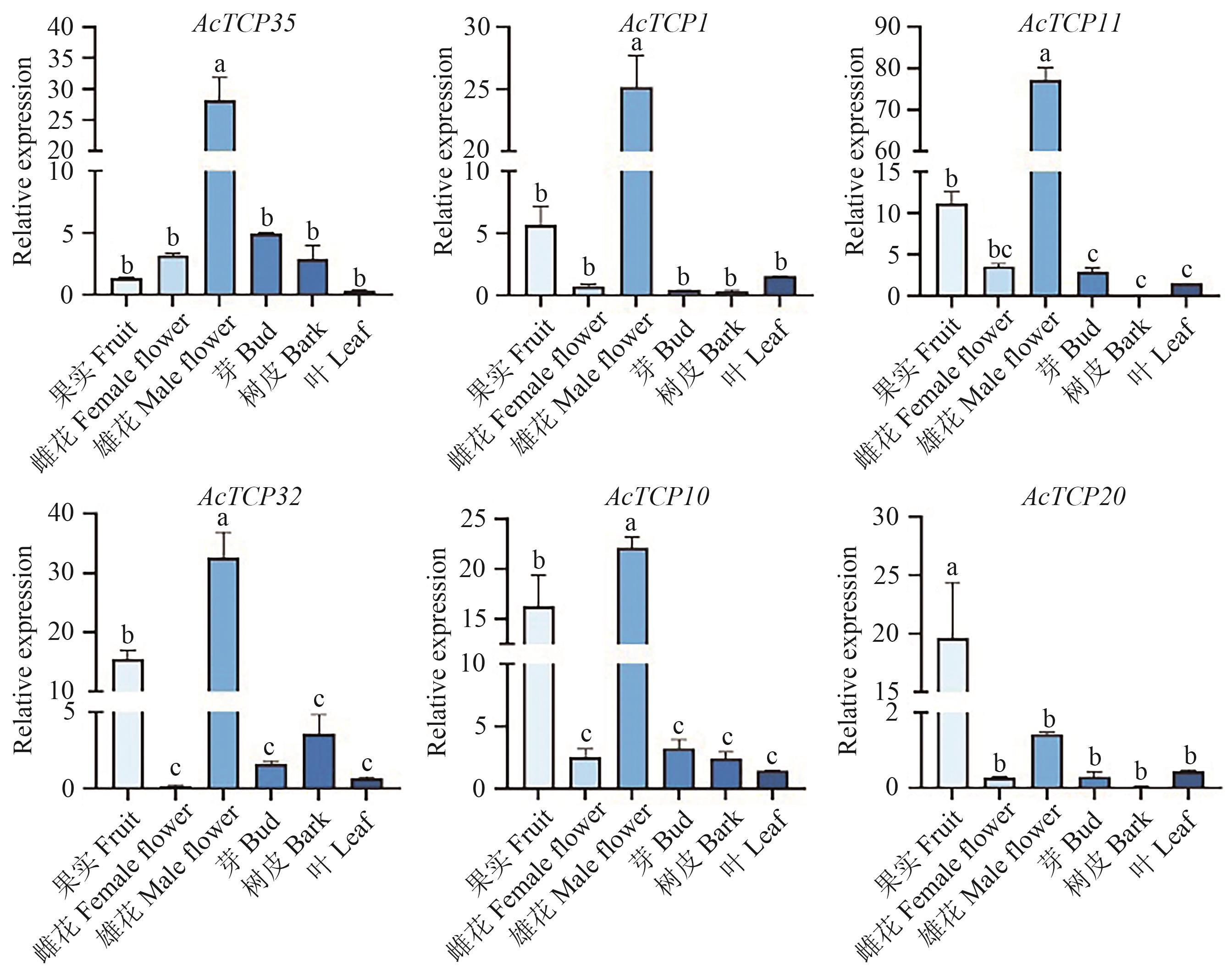
Fig. 6 Analysis of tissue expression pattern of AcTCP genes in A. chinensis var. HongyangDifferent letters indicate significant difference at P<0.05 level. The same below

Fig. 8 Expressions of AcTCP genes in different hormone treatmentsA: ABA treatment; B: ET treatment; C: GA3 treatment; D: CPPU treatment. Horizontal coordinates indicate different treatment times
| 1 | Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize [J]. Nature, 1997, 386(6624): 485-488. |
| 2 | Luo D, Carpenter R, Vincent C, et al. Origin of floral asymmetry in Antirrhinum [J]. Nature, 1996, 383(6603): 794-799. |
| 3 | Kosugi S, Ohashi Y. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene [J]. Plant Cell, 1997, 9(9): 1607-1619. |
| 4 | Cubas P, Lauter N, Doebley J, et al. The TCP domain: a motif found in proteins regulating plant growth and development [J]. Plant J, 1999, 18(2): 215-222. |
| 5 | Ori N, Cohen AR, Etzioni A, et al. Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato [J]. Nat Genet, 2007, 39(6): 787-791. |
| 6 | Aguilar-Martínez JA, Sinha N. Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development [J]. Front Plant Sci, 2013, 4: 406. |
| 7 | Palatnik JF, Allen E, Wu XL, et al. Control of leaf morphogenesis by microRNAs [J]. Nature, 2003, 425(6955): 257-263. |
| 8 | Zhao YF, Broholm SK, Wang F, et al. TCP and MADS-box transcription factor networks regulate heteromorphic flower type identity in Gerbera hybrida [J]. Plant Physiol, 2020, 184(3): 1455-1468. |
| 9 | 陈薇薇. 天然三倍体枇杷花期提前及重瓣花发育的分子调控机制研究 [D]. 重庆: 西南大学, 2020. |
| Chen WW. The molecular regulation mechanisms of early-flowering and double-flower development of natural triploid loquat [D]. Chongqing: Southwest University, 2020. | |
| 10 | Damerval C, Le Guilloux M, Jager M, et al. Diversity and evolution of CYCLOIDEA-like TCP genes in relation to flower development in Papaveraceae [J]. Plant Physiol, 2007, 143(2): 759-772. |
| 11 | Luo D, Carpenter R, Copsey L, et al. Control of organ asymmetry in flowers of Antirrhinum [J]. Cell, 1999, 99(4): 367-376. |
| 12 | 范永明. 芍药雄蕊瓣化关键基因挖掘及功能验证 [D]. 北京: 北京林业大学, 2021. |
| Fan YM. Identification and functional verification of key genes for petaloid stamens in Paeonia lactiflora [D]. Beijing: Beijing Forestry University, 2021. | |
| 13 | Nag A, King S, Jack T. miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis [J]. Proc Natl Acad Sci U S A, 2009, 106(52): 22534-22539. |
| 14 | Takeda T, Amano K, Ohto MA, et al. RNA interference of the Arabidopsis putative transcription factor TCP16 gene results in abortion of early pollen development [J]. Plant Mol Biol, 2006, 61(1/2): 165-177. |
| 15 | Tatematsu K, Nakabayashi K, Kamiya Y, et al. Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana [J]. Plant J, 2008, 53(1): 42-52. |
| 16 | Giraud E, Ng S, Carrie C, et al. TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana [J]. Plant Cell, 2010, 22(12): 3921-3934. |
| 17 | Pruneda-Paz JL, Breton G, Para A, et al. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock [J]. Science, 2009, 323(5920): 1481-1485. |
| 18 | 李晓静, 李圣龙, 岳亮亮. 石榴TCP基因家族的生物信息学与表达分析 [J]. 江苏农业科学, 2024, 52(1): 41-48. |
| Li XJ, Li SL, Yue LL. Bioinformatics and expression analysis of TCP gene family in Punica granatum L [J]. Jiangsu Agric Sci, 2024, 52(1): 41-48. | |
| 19 | Jiu ST, Xu Y, Wang JY, et al. Genome-wide identification, characterization, and transcript analysis of the TCP transcription factors in Vitis vinifera [J]. Front Genet, 2019, 10: 1276. |
| 20 | Edris S, Abulfaraj AA, Makki RM, et al. Early fruit development regulation-related genes concordantly expressed with TCP transcription factors in tomato (Solanum lycopersicum) [J]. Curr Issues Mol Biol, 2023, 45(3): 2372-2380. |
| 21 | Wei W, Hu Y, Cui MY, et al. Identification and transcript analysis of the TCP transcription factors in the diploid woodland strawberry Fragaria vesca [J]. Front Plant Sci, 2016, 7: 1937. |
| 22 | Chen CQ, Zhang Y, Chen YF, et al. Sweet cherry TCP gene family analysis reveals potential functions of PavTCP1, PavTCP2 and PavTCP3 in fruit light responses [J]. BMC Genomics, 2024, 25(1): 3. |
| 23 | 陈延松, 袁华玲, 卫文渊, 等. 夏季遮阳对‘红阳’猕猴桃净光合速率的影响及其与生理生态因子的关系 [J]. 果树学报, 2017, 34(9): 1144-1151. |
| Chen YS, Yuan HL, Wei WY, et al. Impact of summer shading on net photosynthetic rate of Actinidia chinensis 'Hongyang' and its related eco-physiological factors [J]. J Fruit Sci, 2017, 34(9): 1144-1151. | |
| 24 | 刘林娅, 杨那, 罗宇璇, 等. ‘红阳’猕猴桃金属硫蛋白基因AcMT2的克隆、表达分析及功能鉴定 [J] .分子植物育种, 2024. . |
| Liu LY, Yang N, Luo YX, et al. Cloning, expression analysis and functional characterization of the metallothionein gene AcMT2 in Actinidia chinensis var. 'Hongyang' [J]. Mol Plant Breed, 2024. . | |
| 25 | 任东立, 黄亚成, 刘林娅, 等. ‘红阳’猕猴桃蔗糖合成酶(AcSUS1)的克隆与表达分析 [J]. 分子植物育种, 2023. . |
| Ren DL, Huang YC, Liu LY, et al. Cloning and expression analysis of sucrose synthase (AcSUS1) from Actinidia Chinensis var. 'Hongyang' [J]. Mol Plant Breed, 2023. . | |
| 26 | 刘林娅, 杨那, 代玥, 等. 一种大量提取猕猴桃不同组织高质量总RNA的方法 [J]. 江西农业学报, 2020, 32(9): 30-34. |
| Liu LY, Yang N, Dai Y, et al. An efficient method for isolating high-quality total RNA from different tissues of kiwifruit (Actinidia chinensis) [J]. Acta Agric Jiangxi, 2020, 32(9): 30-34. | |
| 27 | Aggarwal P, Das Gupta M, Joseph AP, et al. Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis [J]. Plant Cell, 2010, 22(4): 1174-1189. |
| 28 | Li ZQ, Ouyang YW, Pan XL, et al. TCP transcription factors in pineapple: genome-wide characterization and expression profile analysis during flower and fruit development [J]. Horticulturae, 2023, 9(7): 799. |
| 29 | 周延培. 柑橘TCP家族生物信息学分析及CsTCP1转录因子参与果实成熟的功能验证 [D]. 武汉: 华中农业大学, 2016. |
| Zhou YP. Bioinformatics identification of TCP gene family in citrus and functional verification of CsTCP1 transcription factor involved in fruit Ripening [D]. Wuhan: Huazhong Agricultural University, 2016. | |
| 30 | Guo ZH, Shu WS, Cheng HY, et al. Expression analysis of TCP genes in peach reveals an involvement of PpTCP.A2 in ethylene biosynthesis during fruit ripening [J]. Plant Mol Biol Report, 2018, 36(4): 588-595. |
| 31 | Leng XP, Wei HR, Xu XZ, et al. Genome-wide identification and transcript analysis of TCP transcription factors in grapevine [J]. BMC Genomics, 2019, 20(1): 786. |
| 32 | 谢尹歌. 草莓转录因子FvTCP9在果实成熟中的功能研究 [D]. 杨凌: 西北农林科技大学, 2019. |
| Xie YG. Functional analysis of strawberry (Fragaria vesca) transcription factor FvTCP9 in fruit Ripening [D]. Yangling: Northwest A & F University, 2019. | |
| 33 | Gupta K, Wani SH, Razzaq A, et al. Abscisic acid: role in fruit development and ripening [J]. Front Plant Sci, 2022, 13: 817500. |
| 34 | Du CL, Cai CL, Lu Y, et al. Identification and expression analysis of invertase family genes during grape (Vitis vinifera L.) berry development under CPPU and GA treatment [J]. Mol Genet Genomics, 2023, 298(3): 777-789. |
| 35 | Liu YJ, An JP, Gao N, et al. MdTCP46 interacts with MdABI5 to negatively regulate ABA signalling and drought response in apple [J]. Plant Cell Environ, 2022, 45(11): 3233-3248. |
| 36 | Wen HF, Chen Y, Du H, et al. Genome-wide identification and characterization of the TCP gene family in cucumber (Cucumis sativus L.) and their transcriptional responses to different treatments [J]. Genes, 2020, 11(11): 1379. |
| 37 | Qi XY, Qu YX, Gao R, et al. The heterologous expression of a Chrysanthemum nankingense TCP transcription factor blocks cell division in yeast and Arabidopsis thaliana [J]. Int J Mol Sci, 2019, 20(19): 4848. |
| [1] | ZHANG Yi-xuan, MA Yu, WANG Tong-tong, SHENG Su-ao, SONG Jia-feng, LYU Zhao-yan, ZHU Xiao-biao, HOU Hua-lan. Genome-wide Identification and Expression Profiles of DIR Gene Family in Potato [J]. Biotechnology Bulletin, 2025, 41(3): 123-136. |
| [2] | QIN Yue, YANG Yan, ZHANG Lei, LU Li-li, LI Xian-ping, JIANG Wei. Identification and Comparative Analysis of the StGAox Genes in Diploid and Tetraploid Potatoes [J]. Biotechnology Bulletin, 2025, 41(3): 146-160. |
| [3] | WANG Chen, LIU Guo-mei, CHEN Chang, ZHANG Jin-long, YAO Lin, SUN Xuan, DU Chun-fang. Genome-wide Identification and Expression Analysis of CCDs Family in Brassia rapa L. [J]. Biotechnology Bulletin, 2025, 41(3): 161-170. |
| [4] | HAN Jiang-tao, ZHANG Shuai-bo, QIN Ya-rui, HAN Shuo-yang, ZHANG Ya-kang, WANG Ji-qing, DU Qing-jie, XIAO Huai-juan, LI Meng. Identification of β-amylase Gene Family in Melon and Their Response to Abiotic Stresses [J]. Biotechnology Bulletin, 2025, 41(3): 171-180. |
| [5] | JIA Zi-jian, WANG Bao-qiang, CHEN Li-fei, WANG Yi-zhen, WEI Xiao-hong, ZHAO Ying. Expression Patterns of CHX Gene Family in Quinoa in Response to NO under Saline-alkali Stress [J]. Biotechnology Bulletin, 2025, 41(2): 163-174. |
| [6] | YAN Wei, CHEN Hui-ting, YE Qing, LIU Guang-chao, LIU Xin, HOU Li-xia. Identification of the Grape HCT Gene Family and Their Responses to Low-temperature Stress [J]. Biotechnology Bulletin, 2025, 41(2): 175-186. |
| [7] | KUANG Jian-hua, CHENG Zhi-peng, ZHAO Yong-jing, YANG Jie, CHEN Run-qiao, CHEN Long-qing, HU Hui-zhen. Expression Analysis of the GH3 Gene Family in Nelumbo nucifera underHormonal and Abiotic Stresses [J]. Biotechnology Bulletin, 2025, 41(2): 221-233. |
| [8] | HUANG Ying, YU Wen-jing, LIU Xue-feng, DIAO Gui-ping. Bioinformatics and Expression Pattern Analysis of Glutathione S-transferase in Populus davidiana × P. bolleana [J]. Biotechnology Bulletin, 2025, 41(2): 248-256. |
| [9] | YANG Yong, YUAN Guo-mei, KANG Xiao-xiao, LIU Ya-ming, WANG Dong-sheng, ZHANG Hai-e. Identification and Expression Analysis of Members of the SWEET Gene Family in Chinese Chestnut [J]. Biotechnology Bulletin, 2025, 41(2): 257-269. |
| [10] | LI Ming, LIU Xiang-yu, WANG Yi-na, HE Si-mei, SHA Ben-cai. Cloning and Functional Characterization of 6-OMT Gene Related to Isocorydine Biosynthesis in Dactylicapnos scandens [J]. Biotechnology Bulletin, 2025, 41(2): 309-320. |
| [11] | DU Pin-ting, WU Guo-jiang, WANG Zhen-guo, LI Yan, ZHOU Wei, ZHOU Ya-xing. Identification and Expression Analysis of CPP Gene Family in Sorghum [J]. Biotechnology Bulletin, 2025, 41(1): 132-142. |
| [12] | HE Cai-lin, LU Jing, GUO Hui-hui, LI Xiao-an, WU Qi. Genome-wide Identification and Expression Analysis of the MADS-box Gene Family in Quinoa [J]. Biotechnology Bulletin, 2025, 41(1): 157-172. |
| [13] | WANG Zi-ao, TIAN Rui, CUI Yong-mei, BAI Yi-xiong, YAO Xiao-hua, AN Li-kun, WU Kun-lun. Bioinformatics and Expression Pattern Analysis of HvnJAZ4 Gene in Hulless Barley [J]. Biotechnology Bulletin, 2025, 41(1): 173-185. |
| [14] | KONG Qing-yang, ZHANG Xiao-long, LI Na, ZHANG Chen-jie, ZHANG Xue-yun, YU Chao, ZHANG Qi-xiang, LUO Le. Identification and Expression Analysis of GRAS Transcription Factor Family in Rosa persica [J]. Biotechnology Bulletin, 2025, 41(1): 210-220. |
| [15] | SONG Bing-fang, LIU Ning, CHENG Xin-yan, XU Xiao-bin, TIAN Wen-mao, GAO Yue, BI Yang, WANG Yi. Identification of Potato G6PDH Gene Family and Its Expression Analysis in Damaged Tubers [J]. Biotechnology Bulletin, 2024, 40(9): 104-112. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||