








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (4): 134-144.doi: 10.13560/j.cnki.biotech.bull.1985.2024-1017
YANG Chao-jie( ), ZHANG Lan, CHEN Hong, HUANG Juan, SHI Tao-xiong, ZHU Li-wei, CHEN Qing-fu, LI Hong-you(
), ZHANG Lan, CHEN Hong, HUANG Juan, SHI Tao-xiong, ZHU Li-wei, CHEN Qing-fu, LI Hong-you( ), DENG Jiao(
), DENG Jiao( )
)
Received:2024-10-21
Online:2025-04-26
Published:2025-04-25
Contact:
LI Hong-you, DENG Jiao
E-mail:15685965728m@sina.cn;lihongyouluod@163.com;ddj613@163.com
YANG Chao-jie, ZHANG Lan, CHEN Hong, HUANG Juan, SHI Tao-xiong, ZHU Li-wei, CHEN Qing-fu, LI Hong-you, DENG Jiao. Functional Identification of the Transcription Factor Gene FtbHLH3 in Regulating Flavonoid Biosynthesis in Fagopyrum tataricum[J]. Biotechnology Bulletin, 2025, 41(4): 134-144.
| 引物名称 Primer name | 引物序列 Primer sequences(5′-3′) | 用途 Purpose |
|---|---|---|
| FtbHLH3-F | 遗传转化 | |
| FtbHLH3-R | ||
| AtCHS | F:AATGGTGATGGCTGGTGCTT;R:TGGTGATGCGGAAGTAGTAGTC | 实时荧光定量 |
| AtCHI | F:GCCTCCTCCAATCCATTATTCC;R:CCTTCCACTTGACAGATAGAGAA | |
| AtF3H | F:CATCGTCTCTAGTCACCTCCAG;R:CTCACTATACTCCTCCGTCACTT | |
| AtDFR | F:CGCCAAGACGCTACTCACT;R:CGGCTTTATCACTTCGTTCTCA | |
| AtANS | F:GTGATTACATAGAAGCAACGAGTG;R:CTAAACCTAGACCGACAGAGAGA | |
| AtANR | F:CGGCGATACAAGGAGTGA;R:AAGGCTTCTCCTCTGTGA | |
| AtUFGT | F:CAACTGGTTTTCCGTTTCTGGTT;R:GCTTCCTCGACGGTTGATACAC | |
| AtFLS | F:TCACAACATTCCGAGGTCCAA;R:CTTCGTCGGGATCGCTTAGA | |
| AtActin2 | F:TGCTGGATTCTGGTGATGGT;R:AAGGTCAAGACGGAGGATGG |
Table 1 Primer sequences used in the study
| 引物名称 Primer name | 引物序列 Primer sequences(5′-3′) | 用途 Purpose |
|---|---|---|
| FtbHLH3-F | 遗传转化 | |
| FtbHLH3-R | ||
| AtCHS | F:AATGGTGATGGCTGGTGCTT;R:TGGTGATGCGGAAGTAGTAGTC | 实时荧光定量 |
| AtCHI | F:GCCTCCTCCAATCCATTATTCC;R:CCTTCCACTTGACAGATAGAGAA | |
| AtF3H | F:CATCGTCTCTAGTCACCTCCAG;R:CTCACTATACTCCTCCGTCACTT | |
| AtDFR | F:CGCCAAGACGCTACTCACT;R:CGGCTTTATCACTTCGTTCTCA | |
| AtANS | F:GTGATTACATAGAAGCAACGAGTG;R:CTAAACCTAGACCGACAGAGAGA | |
| AtANR | F:CGGCGATACAAGGAGTGA;R:AAGGCTTCTCCTCTGTGA | |
| AtUFGT | F:CAACTGGTTTTCCGTTTCTGGTT;R:GCTTCCTCGACGGTTGATACAC | |
| AtFLS | F:TCACAACATTCCGAGGTCCAA;R:CTTCGTCGGGATCGCTTAGA | |
| AtActin2 | F:TGCTGGATTCTGGTGATGGT;R:AAGGTCAAGACGGAGGATGG |
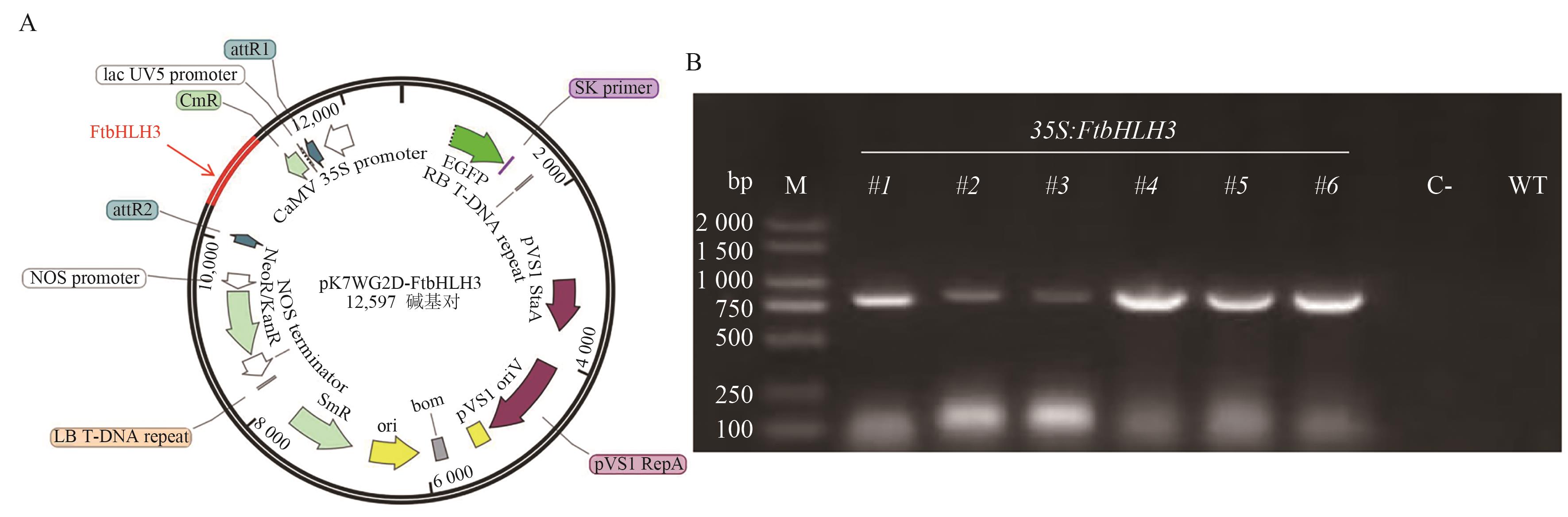
Fig. 3 Overexpression vector pK7WG2D-FtbHLH3 (A) and identification of positive seedlings of transgenic Arabidopsis (B)A: Overexpression vector pK7WG2D-FtbHLH3; B: FtbHLH3 complements the T1 generation seedlings of Arabidopsistt8 mutant. M: DL 2 000 marker; 1-6: the gDNA of each transgenic plant used as a template; C-: negative control (using water as template); WT: negative control (using wild-type gDNA as template)

Fig. 4 Seedling analysis of FtbHLH3 transgenic Arabidopsis thalianaattt8 mutantA: Phenotypic observation of seedlings of WT, attt8 mutant, and 35S:FtbHLH3 line. B: DMACA staining of seedlings and microscope observation (40×). C: Determination of anthocyanin content. D: Determination of PAs content. E: Determination of total flavonoid content. F: RT-qPCR analysis. WT: Wild type; tt8: Arabidopsis attt8 mutant; 35S: FtbHLH3: FtbHLH3 complements the Arabidopsisattt8 mutant; *P<0.05; **P<0.01; ***P<0.001. The same below

Fig. 5 Seed analysis of FtbHLH3 transgenic Arabidopsis thalianaattt8 mutantA: Observation of seed phenotypes of WT, attt8 mutant, and 35S:FtbHLH3 line. B: DMACA staining and microscope observation (40×). C: Determination of anthocyanin content. D: Determination of PAs content. E: Determination of total flavonoid content. F: RT-qPCR analysis
| 1 | 杨学乐, 张璐, 李志清, 等. 苦荞种质资源表型性状的遗传多样性分析 [J]. 作物杂志, 2020(5): 53-58. |
| Yang XL, Zhang L, Li ZQ, et al. Diversity analysis of Tartary buckwheat germplasms based on phenotypic traits [J]. Crops, 2020(5): 53-58. | |
| 2 | 彭艳, 江燕, 刘代铃, 等. 有机质和微生物菌剂对苦荞连作农艺性状及土壤酶活性的影响 [J]. 分子植物育种, 2023,21(4): 1287-1293. |
| Peng Y, Jiang Y, Liu DL, et al. Mitigation of organic matter and microbial inoculants on continuous cropping obstacle of Tartary buckwheat [J]. Molecular Plant Breeding, 2023,21(4): 1287-1293. | |
| 3 | 姚鑫, 刘婷婷, 阮景军, 等. H2O2浸种对苦荞幼苗根系生长和抗氧化酶活性的影响 [J]. 分子植物育种, 2024, 22(7): 2354-2360. |
| Yao X, Liu TT, Ruan JJ, et al. Effects of H2O2 seed soaking on root growth and antioxidant enzyme activities of Tartary buckwheat(F. tataricum)seedlings [J]. Mol Plant Breed, 2024, 22(7): 2354-2360. | |
| 4 | 严泽, 杨玉婷, 巴宗, 等. 西藏昌都市八宿县不同苦荞品种营养成分检测分析 [J]. 西藏农业科技, 2023, 45(1): 10-13. |
| Yan Z, Yang YT, Ba Z, et al. Analysis of nutrient composition from different Tartary buckwheat varieties in Basu County, Changdu City, Xizang autonomous region [J]. Tibet J Agric Sci, 2023, 45(1): 10-13. | |
| 5 | Hou ZX, Hu YY, Yang XB, et al. Antihypertensive effects of Tartary buckwheat flavonoids by improvement of vascular insulin sensitivity in spontaneously hypertensive rats [J]. Food Funct, 2017, 8(11): 4217-4228. |
| 6 | Hwang D, Kang MJ, Kang CW, et al. Kaempferol-3-O-β-rutinoside suppresses the inflammatory responses in lipopolysaccharide-stimulated RAW264.7 cells via the NF-κB and MAPK pathways [J]. Int J Mol Med, 2019, 44(6): 2321-2328. |
| 7 | Zhou XL, Chen ZD, Zhou YM, et al. The effect of Tartary buckwheat flavonoids in inhibiting the proliferation of MGC80-3 cells during seed germination [J]. Molecules, 2019, 24(17): 3092. |
| 8 | Dzah CS, Duan YQ, Zhang HH, et al. Ultrasound-, subcritical water- and ultrasound assisted subcritical water-derived Tartary buckwheat polyphenols show superior antioxidant activity and cytotoxicity in human liver carcinoma cells [J]. Food Res Int, 2020, 137: 109598. |
| 9 | Cao P, Wu Y, Li YP, et al. The important role of glycerophospholipid metabolism in the protective effects of polyphenol-enriched Tartary buckwheat extract against alcoholic liver disease [J]. Food Funct, 2022, 13(20): 10415-10425. |
| 10 | 贾赵东, 马佩勇, 边小峰, 等. 植物花青素合成代谢途径及其分子调控 [J]. 西北植物学报, 2014, 34(7): 1496-1506. |
| Jia ZD, Ma PY, Bian XF, et al. Biosynthesis metabolic pathway and molecular regulation of plants anthocyanin [J]. Acta Bot Boreali Occidentalia Sin, 2014, 34(7): 1496-1506. | |
| 11 | Pourcel L, Routaboul JM, Kerhoas L, et al. TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat [J]. Plant Cell, 2005, 17(11): 2966-2980. |
| 12 | Zhao MY, Jin JY, Gao T, et al. Glucosyltransferase CsUGT78A14 regulates flavonols accumulation and reactive oxygen species scavenging in response to cold stress in Camellia sinensis [J]. Front Plant Sci, 2019, 10: 1675. |
| 13 | Chandler VL, Radicella JP, Robbins TP, et al. Two regulatory genes of the maize anthocyanin pathway are homologous: isolation of B utilizing R genomic sequences [J]. Plant Cell, 1989, 1(12): 1175-1183. |
| 14 | 赖瑞联, 吴如健, 赖钟雄. 植物类黄酮的分类、药理活性及其生物合成调控研究进展 [J]. 东南园艺, 2024, 12(2): 81-99. |
| Lai RL, Wu RJ, Lai ZX. Research progress on the classification, pharmacological activity, and biosynthesis regulation of plant flavonoid [J]. Southeast Hortic, 2024, 12(2): 81-99. | |
| 15 | Deng J, Li JJ, Su MY, et al. A bHLH gene NnTT8 of Nelumbo nucifera regulates anthocyanin biosynthesis [J]. Plant Physiol Biochem, 2021, 158: 518-523. |
| 16 | Hu R, Zhu MC, Chen S, et al. BnbHLH92a negatively regulates anthocyanin and proanthocyanidin biosynthesis in Brassica napus [J]. Crop J, 2023, 11(2): 374-385. |
| 17 | Li XJ, Cao LJ, Jiao BB, et al. The bHLH transcription factor AcB2 regulates anthocyanin biosynthesis in onion (Allium cepa L.) [J]. Hortic Res, 2022, 9: uhac128. |
| 18 | Zhao R, Song XX, Yang N, et al. Expression of the subgroup IIIf bHLH transcription factor CpbHLH1 from Chimonanthus praecox (L.) in transgenic model plants inhibits anthocyanin accumulation [J]. Plant Cell Rep, 2020, 39(7): 891-907. |
| 19 | Wang FB, Zhu H, Kong WL, et al. The Antirrhinum AmDEL gene enhances flavonoids accumulation and salt and drought tolerance in transgenic Arabidopsis [J]. Planta, 2016, 244(1): 59-73. |
| 20 | 王丽娟. 苦荞FtTT8基因的克隆及调控花色苷和原花青素生物合成的功能验证 [D]. 贵阳: 贵州师范大学, 2023. |
| Wang LJ. Cloning of Fagopyrum tataricum FtTT8 gene and functional verification of regulating anthocyanin and proanthocyanidins biosynthesis [D]. Guiyang: Guizhou Normal University, 2023. | |
| 21 | Bai YC, Li CL, Zhang JW, et al. Characterization of two Tartary buckwheat R2R3-MYB transcription factors and their regulation of proanthocyanidin biosynthesis [J]. Physiol Plant, 2014, 152(3): 431-440. |
| 22 | Zhang D, Jiang CL, Huang CH, et al. The light-induced transcription factor FtMYB116 promotes accumulation of rutin in Fagopyrum tataricum [J]. Plant Cell Environ, 2019, 42(4): 1340-1351. |
| 23 | Zhang KX, Logacheva MD, Meng Y, et al. Jasmonate-responsive MYB factors spatially repress rutin biosynthesis in Fagopyrum tataricum [J]. J Exp Bot, 2018, 69(8): 1955-1966. |
| 24 | 姚攀锋. 苦荞WD40转录因子的基因克隆及其对花青素合成的影响 [D]. 雅安: 四川农业大学, 2016. |
| Yao PF. Gene cloning of WD40 transcription factor from Tartary buckwheat and its effect on anthocyanin synthesis [D]. Ya'an: Sichuan Agricultural University, 2016. | |
| 25 | 牛毅男. 水分胁迫对苦荞次生代谢调控机制初步探究 [D]. 杨凌: 西北农林科技大学, 2020. |
| Niu YN. Preliminary study on the regulation mechanism of water stress on secondary metabolism of Tartary buckwheat [D]. Yangling: Northwest A & F University, 2020. | |
| 26 | Li HY, Lv QY, Ma C, et al. Metabolite profiling and transcriptome analyses provide insights into the flavonoid biosynthesis in the developing seed of Tartary buckwheat (Fagopyrum tataricum) [J]. J Agric Food Chem, 2019, 67(40): 11262-11276. |
| 27 | 吕秋谕, 孙培媛, 冉彬, 等. 苦荞转录因子基因FtbHLH3的克隆、亚细胞定位及表达分析 [J]. 生物技术通报, 2023, 39(8): 194-203. |
| Lv QY, Sun PY, Ran B, et al. Cloning, subcellular localization and expression analysis of the transcription factor gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnol Bull, 2023, 39(8): 194-203. | |
| 28 | Liu RJ, Wang YY, Tang S, et al. Genome-wide identification of the tea plant bHLH transcription factor family and discovery of candidate regulators of trichome formation [J]. Sci Rep, 2021, 11(1): 10764. |
| 29 | 高建强, 梁华, 赵军. 植物遗传转化农杆菌浸花法研究进展 [J]. 中国农学通报, 2010, 26(16): 22-25. |
| Gao JQ, Liang H, Zhao J. Progress on the floral-dip method of agribacterium-mediated plant transformation [J]. Chin Agric Sci Bull, 2010, 26(16): 22-25. | |
| 30 | Chowdhury J, Ferdous J, Lihavainen J, et al. Fluorogenic properties of 4-dimethylaminocinnamaldehyde (DMACA) enable high resolution imaging of cell-wall-bound proanthocyanidins in plant root tissues [J]. Front Plant Sci, 2023, 13: 1060804. |
| 31 | 田建华. 沙棘原花青素测定方法学考察及含量测定 [J]. 山西林业科技, 2021, 50(4): 11-13. |
| Tian JH. Methodological inspection and content determination of proanthocyanidins in Hippophae rhamnoides [J]. Shanxi For Sci Technol, 2021, 50(4): 11-13. | |
| 32 | 孙小倩. 苦荞转录因子FtMYBF的克隆及功能分析 [D]. 贵阳: 贵州师范大学, 2022. |
| Sun XQ. Cloning and functional analysis of buckwheat transcription factor FtMYBF [D]. Guiyang: Guizhou Normal University, 2022. | |
| 33 | 张凯敏, 耿贵工, 乔枫. 枸杞果实发育期酶活性、基因表达与类黄酮积累的相关性分析 [J]. 分子植物育种,2024,22(22): 7515-7524. |
| Zhang KM, Gen GG, Qiao F. Correlation analysis of enzyme activity, gene expression and flavonoid accumulation during fruit development of Lycium chinense [J]. Molecular Plant Breeding,2024,22(22): 7515-7524. | |
| 34 | Li W, Wang B, Wang M, et al. Cloning and characterization of a potato StAN11 gene involved in anthocyanin biosynthesis regulation [J]. J Integr Plant Biol, 2014, 56(4): 364-372. |
| 35 | 胡锦锦, 李素贞, 马旭辉, 等. 玉米花青素生物合成代谢调控 [J]. 生物技术通报, 2024, 40(6): 34-44. |
| Hu JJ, Li SZ, Ma XH, et al. Regulation of maize anthocyanin biosynthesis metabolism [J]. Biotechnol Bull, 2024, 40(6): 34-44. | |
| 36 | Li PH, Chen BB, Zhang GY, et al. Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bHLH transcription factor MtTT8 [J]. New Phytol, 2016, 210(3): 905-921. |
| 37 | Zhao PC, Li XX, Jia JT, et al. bHLH92 from sheepgrass acts as a negative regulator of anthocyanin/proanthocyandin accumulation and influences seed dormancy [J]. J Exp Bot, 2019, 70(1): 269-284. |
| 38 | Tao RY, Yu WJ, Gao YH, et al. Light-induced basic/helix-loop-Helix64 enhances anthocyanin biosynthesis and undergoes CONSTITUTIVELY PHOTOMORPHOGENIC1-mediated degradation in pear [J]. Plant Physiol, 2020, 184(4): 1684-1701. |
| [1] | PENG Ting, LIN Ying, TAN Yuan-yuan, RAO Ying, HUANG Qin, ZHANG Wen-e, WANG Bo, TIAN Rui-feng, LIU Guo-feng. Cloning and Expression Analysis of AwANS Genes in Allium wallichii [J]. Biotechnology Bulletin, 2025, 41(3): 230-239. |
| [2] | WANG Bin, WANG Yu-kun, XIAO Yan-hui. Comparative Transcriptomic Analysis of Clove Basil (Ocimum gratissimum) Leaves in Response to Cadmium Stress [J]. Biotechnology Bulletin, 2025, 41(3): 255-270. |
| [3] | YANG Yong, CAO Rui, KANG Xiao-xiao, LIU Jing, WANG Xuan, ZHANG Hai-e. Identification and Expression Analysis of 13 Gene Families in the Chestnut Flavonoid Synthesis Pathway [J]. Biotechnology Bulletin, 2025, 41(2): 270-283. |
| [4] | SONG Qian-na, DUAN Yong-hong, FENG Rui-yun. Establishment of CRISPR/Cas9-mediated Highly Efficient Gene Editing System in Microtubers of Potatoes [J]. Biotechnology Bulletin, 2024, 40(9): 33-41. |
| [5] | LI Yu-qing, WU Nan, LUO Jian-rang. Cloning and Functional Analysis of bHLH Gene Related to Anthocyanin Synthesis in Paeonia qiui [J]. Biotechnology Bulletin, 2024, 40(8): 174-185. |
| [6] | ZHANG Mei-yu, ZHAO Yu-bin, WANG Ling-yun, SONG Yuan-da, ZHAO Xin-he, REN Xiao-jie. Research Progress in the Production of Functional Fatty Acid DHA by Microalga Thraustochytrids [J]. Biotechnology Bulletin, 2024, 40(6): 81-94. |
| [7] | MEI Xian-jun, SONG Hui-yang, LI Jing-hao, MEI Chao, SONG Qian-na, FENG Rui-yun, CHEN Xi-ming. Cloning and Expression Analysis of StDof5 Gene in Potato [J]. Biotechnology Bulletin, 2024, 40(3): 181-192. |
| [8] | YUAN Xin-yu, ZHONG Cai-hong, ZHANG Long, ZHENG Hao, LI Ji-tao, ZHANG Qiong. Identification and Function Analysis of AcMYB88 in Kiwifruit (Actinidia chinensis) [J]. Biotechnology Bulletin, 2024, 40(2): 183-196. |
| [9] | LI Can, JIANG Xiang-ning, GAI Ying. Cloning of the LkF3H2 Gene in Larix kaempferi and Its Function in Regulating Flavonoid Metabolism [J]. Biotechnology Bulletin, 2024, 40(2): 245-252. |
| [10] | HUA Xuan, TIAN Bo-wen, ZHOU Xin-tong, JIANG Zi-han, WANG Shi-qi, HUANG Qian-hui, ZHANG Jian, CHEN Yan-hong. Cloning SmERF B3-45 from Salix matsudana and Functional Analysis on Its Tolerance to Salt [J]. Biotechnology Bulletin, 2024, 40(12): 124-135. |
| [11] | HAN Le-le, SONG Wen-di, BIAN Jia-shen, LI Yang, YANG Shuang-sheng, CHEN Zi-yi, LI Xiao-wei. Revealing the Flavonoid Biosynthesis of Soybean GmERD15c under Salt Stress by Combined Analysis of Transcriptome and Metabolome [J]. Biotechnology Bulletin, 2024, 40(10): 243-252. |
| [12] | WANG Bin, YUAN Xiao, JIANG Yuan-yuan, WANG Yu-kun, XIAO Yan-hui, HE Jin-ming. Cloning of bHLH96 Gene and Its Roles in Regulating the Biosynthesis of Peppermint Terpenes [J]. Biotechnology Bulletin, 2024, 40(1): 281-293. |
| [13] | LYU Qiu-yu, SUN Pei-yuan, RAN Bin, WANG Jia-rui, CHEN Qing-fu, LI Hong-you. Cloning, Subcellular Localization and Expression Analysis of the Transcription Factor Gene FtbHLH3 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 194-203. |
| [14] | WANG Jia-rui, SUN Pei-yuan, KE Jin, RAN Bin, LI Hong-you. Cloning and Expression Analyses of C-glycosyltransferase Gene FtUGT143 in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2023, 39(8): 204-212. |
| [15] | LIU Hui, LU Yang, YE Xi-miao, ZHOU Shuai, LI Jun, TANG Jian-bo, CHEN En-fa. Comparative Transcriptome Analysis of Cadmium Stress Response Induced by Exogenous Sulfur in Tartary Buckwheat [J]. Biotechnology Bulletin, 2023, 39(5): 177-191. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||