








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (5): 310-319.doi: 10.13560/j.cnki.biotech.bull.1985.2024-0992
QU Shan1( ), ZHAO Yue1, LI Ya-hua1, ZHENG Gui-ling2, XIAN Hong-quan1(
), ZHAO Yue1, LI Ya-hua1, ZHENG Gui-ling2, XIAN Hong-quan1( )
)
Received:2024-10-12
Online:2025-05-26
Published:2025-06-05
Contact:
XIAN Hong-quan
E-mail:raymond0416@163.com;hqxian00517@163.com
QU Shan, ZHAO Yue, LI Ya-hua, ZHENG Gui-ling, XIAN Hong-quan. A Study on the Interaction between Transcriptional Factor and Protein of Tachi2 Chitinase Gene in Trichoderma asperellum[J]. Biotechnology Bulletin, 2025, 41(5): 310-319.
| 引物名称 Primer name | 序列 Sequence (5′-3′) | 限制性内切酶 Restriction enzyme | 用途 Usage |
|---|---|---|---|
| 47S | CCG | EcoR Ⅰ | 点对点验证 |
| 47A | CG | BamH Ⅰ | 点对点验证 |
| H63S | TCCC | Xma I | 点对点验证 |
| H63A | CG | BamH Ⅰ | 点对点验证 |
| 5′AD | CTATTCGATGATGAAGATACCCCACCAAACCC | - | 点对点验证 |
| 3′AD | AGTGAACTTGGGGGGTTTTTCAGTATCTACGAT | - | 点对点验证 |
| H63S-B/N | CG | BamH Ⅰ | GST pull-down |
| H63A-B/N | ATTT | Not I | GST pull-down |
| CE63S | CG | BamH I | BiFC |
| CE63A | GG | Kpn I | BiFC |
| NE47S | CG | BamH I | BiFC |
| NE47A | GG | Kpn I | BiFC |
Table 1 Primers used in the experiment
| 引物名称 Primer name | 序列 Sequence (5′-3′) | 限制性内切酶 Restriction enzyme | 用途 Usage |
|---|---|---|---|
| 47S | CCG | EcoR Ⅰ | 点对点验证 |
| 47A | CG | BamH Ⅰ | 点对点验证 |
| H63S | TCCC | Xma I | 点对点验证 |
| H63A | CG | BamH Ⅰ | 点对点验证 |
| 5′AD | CTATTCGATGATGAAGATACCCCACCAAACCC | - | 点对点验证 |
| 3′AD | AGTGAACTTGGGGGGTTTTTCAGTATCTACGAT | - | 点对点验证 |
| H63S-B/N | CG | BamH Ⅰ | GST pull-down |
| H63A-B/N | ATTT | Not I | GST pull-down |
| CE63S | CG | BamH I | BiFC |
| CE63A | GG | Kpn I | BiFC |
| NE47S | CG | BamH I | BiFC |
| NE47A | GG | Kpn I | BiFC |
| 组别 Group | 蛋白 Protein |
|---|---|
| 实验组1 | GST-H63蛋白+His-47蛋白 |
| 实验组2 | GST标签蛋白+His-47蛋白 |
| 实验组3 | GST标签蛋白 |
| 对照组1 | 纯化的His-47蛋白 |
| 对照组2 | 纯化的GST-H63蛋白 |
Table 2 GST pull-down experimental design
| 组别 Group | 蛋白 Protein |
|---|---|
| 实验组1 | GST-H63蛋白+His-47蛋白 |
| 实验组2 | GST标签蛋白+His-47蛋白 |
| 实验组3 | GST标签蛋白 |
| 对照组1 | 纯化的His-47蛋白 |
| 对照组2 | 纯化的GST-H63蛋白 |
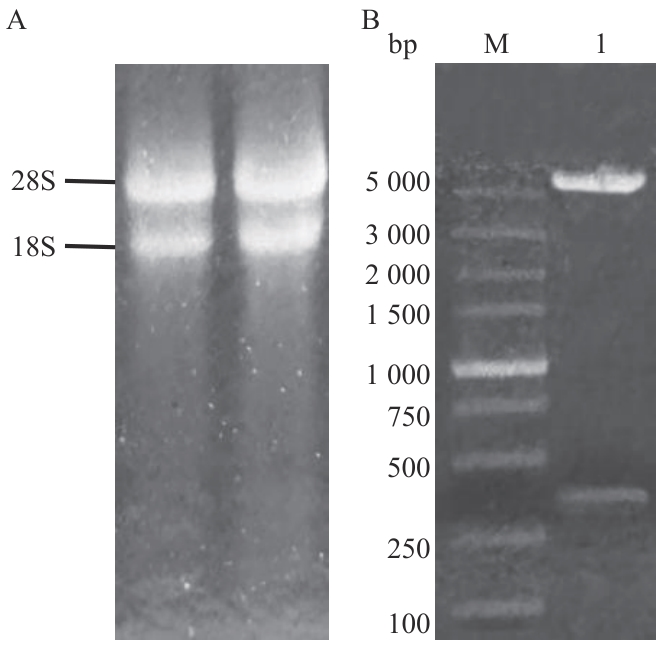
Fig. 1 pGBKT7-47 vector constructionA: RNA of Trichoderma asperellum; B: PGBKT7-47 double enzyme digestion verification (pGBKT7-47 double enzyme-digested product; M: 5000 bp DNA marker)
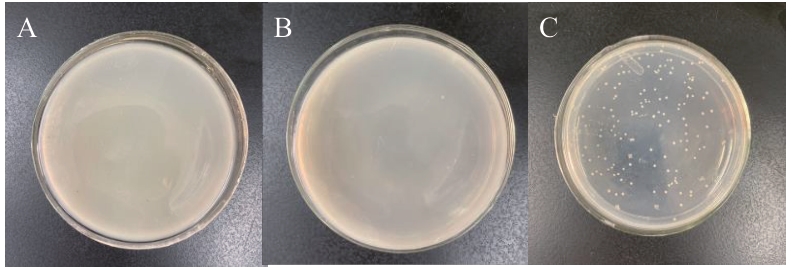
Fig. 2 pGADT7-47 vector self-activation and toxicity testA: Y2H Gold yeast transformant of pGBKT7-47 and pGADT7; B: Y2H Gold yeast transformant of pGADT7-T and pGBKT7-lam; C: Y2H Gold yeast transformant of pGADT7-T and pGBKT7-53
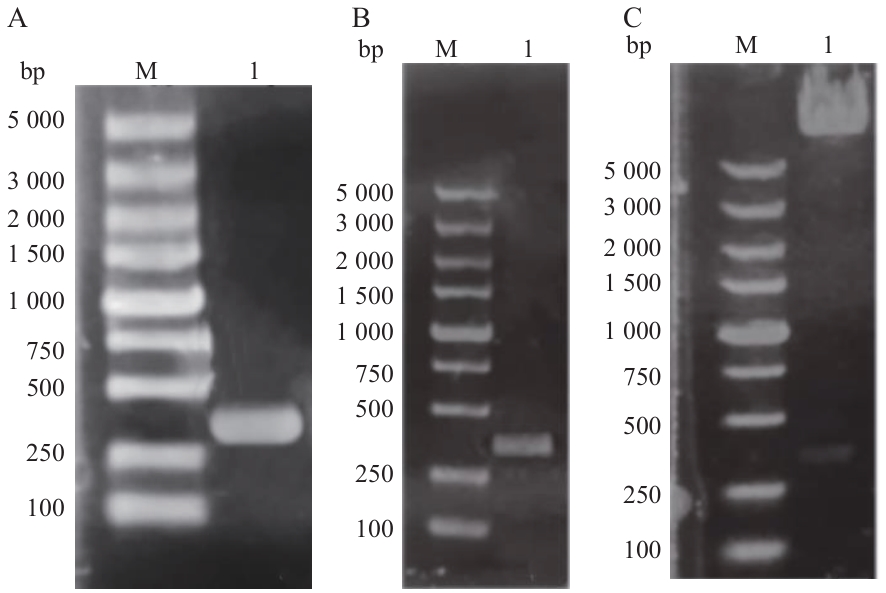
Fig. 3 Construction of interacting pGADT7-H63 vectorA: H63 gene PCR product; B: H63 bacterial liquid PCR product validation; C: PGADT7-63 double enzyme digestion product; M: DL5000 bp DNA marker
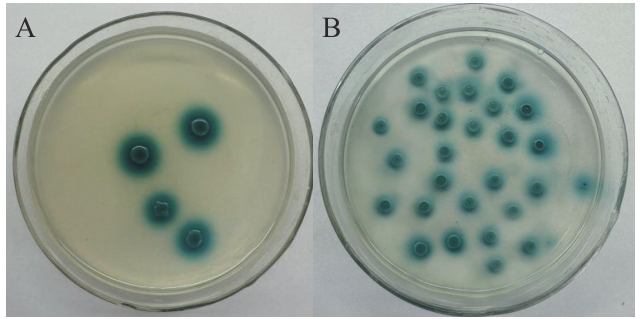
Fig. 4 Verification of the interaction of pGBKT7-47 with pGADT7-H63 through yeast two-hybridA: Y2H Gold yeast transformant of pGBKT7-47 and pGADT7-H63; B: Y2H Gold yeast transformant of pGADT7-T and pGBKT7-53
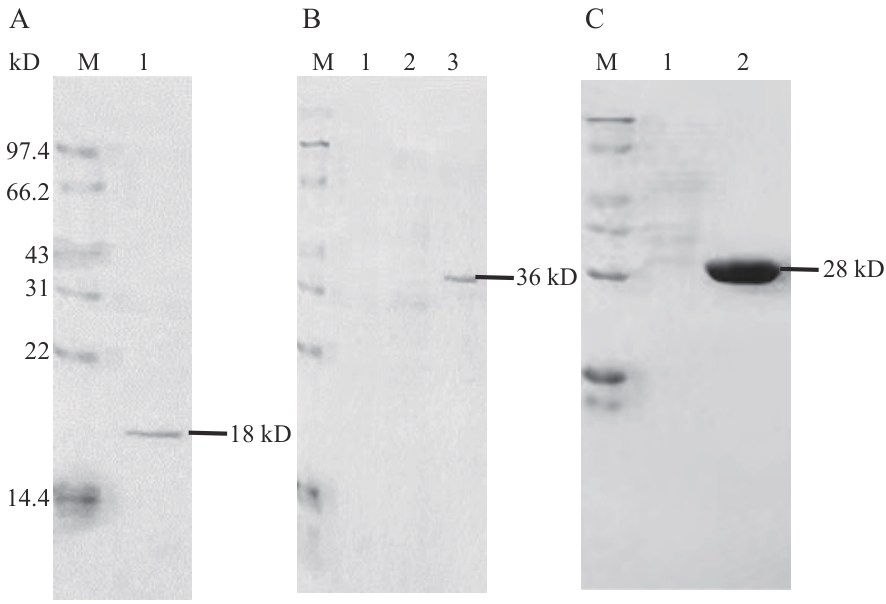
Fig. 6 SDS-PAGE electrophoresis map of recombinant proteinA: Purified 47 protein; B: purified pGEX4T-1-H63 protein; C: purified pGEX4T-1 protein; M: standard protein marker
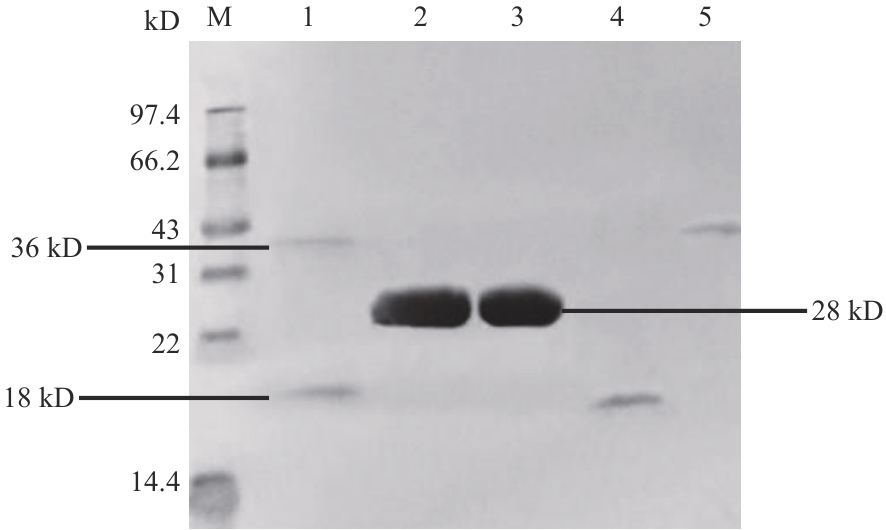
Fig. 7 Protein interaction validation of GST-H63 and His-47 in vitro1: GST-H63 protein and His-47 protein; 2: His-47 protein and GST protein; 3: GST; 4: His-47 protein; 5: GST-H63 protein; M: standard protein marker
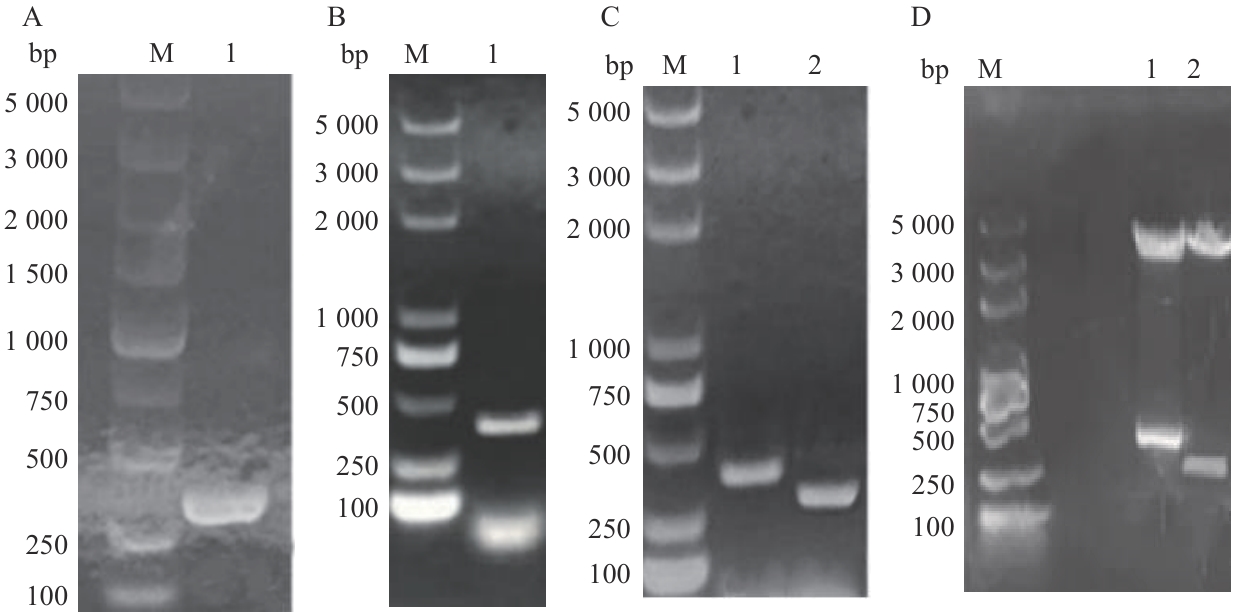
Fig. 8 Construction of BiFC experimental carrierA: H63 gene PCR product; B: PCR product of transcription factor gene 47; C: PCR validation of bacterial liquid (1: PCR product of DH5 α/B-T1-47 bacterial liquid; 2: PCR product of DH5 α/B-T1-H63 bacterial liquid); D: double enzyme digestion validation (1: NE-47 double enzyme cleavage product; 2: CE-63 double enzyme cleavage product); M: DL5000bp DNA marker
| 1 | Sharon E, Chet I, Spiegel Y. Trichoderma as a biological control agent [M]//Davies K, Spiegel Y, eds. Biological Control of Plant-Parasitic Nematodes. Dordrecht: Springer Netherlands, 2011: 183-201. |
| 2 | Mukherjee PK, Buensanteai N, Moran-Diez ME, et al. Functional analysis of non-ribosomal peptide synthetases (NRPSs) in Trichoderma virens reveals a polyketide synthase (PKS)/NRPS hybrid enzyme involved in the induced systemic resistance response in maize [J]. Microbiology, 2012, 158(Pt 1): 155-165. |
| 3 | Woo SL, Hermosa R, Lorito M, et al. Trichoderma: a multipurpose, plant-beneficial microorganism for eco-sustainable agriculture [J]. Nat Rev Microbiol, 2023, 21(5): 312-326. |
| 4 | Kappel L, Münsterkötter M, Sipos G, et al. Chitin and chitosan remodeling defines vegetative development and Trichoderma biocontrol [J]. PLoS Pathog, 2020, 16(2): e1008320. |
| 5 | Bae SJ, Park YH, Bae HJ, et al. Molecular identification, enzyme assay, and metabolic profiling of Trichoderma spp [J]. J Microbiol Biotechnol, 2017, 27(6): 1157-1162. |
| 6 | Al-Ani LKT. A patent survey of Trichoderma spp. (from 2007 to 2017) [M]//Intellectual Property Issues in Microbiology. Singapore: Springer Singapore, 2019: 163-192. |
| 7 | Manzar N, Kashyap AS, Goutam RS, et al. Trichoderma: advent of versatile biocontrol agent, its secrets and insights into mechanism of biocontrol potential [J]. Sustainability, 2022, 14(19): 12786. |
| 8 | 薛德星, 李美, 高兴祥, 等. 生防菌棘孢木霉的分离鉴定及生物学特性研究 [J]. 山东农业科学, 2023, 55(10): 118-123. |
| Xue DX, Li M, Gao XX, et al. Isolation, identification and biological characteristics of Trichoderma asperellum GT30 [J]. Shandong Agric Sci, 2023, 55(10): 118-123. | |
| 9 | 廉华, 马光恕, 李梅, 等. 棘孢木霉菌剂对黄瓜生理特性及产质量的影响 [J]. 中国农业大学学报, 2021, 26(6): 42-52. |
| Lian H, Ma GS, Li M, et al. Effects of Trichoderma asperellum agents on physiological characteristics, yield and quality of cucumber [J]. J China Agric Univ, 2021, 26(6): 42-52. | |
| 10 | Di Rosa M, Distefano G, Zorena K, et al. Chitinases and immunity: Ancestral molecules with new functions [J]. Immunobiology, 2016, 221(3): 399-411. |
| 11 | Dukare AS, Paul S, Asha AD, et al. Role of bacterial and fungal chitinases in integrated management of pest and diseases of agro-horticultural crops [M]//Microbes for Sustainable lnsect Pest Management. Cham: Springer International Publishing, 2021: 33-57. |
| 12 | 卢传琦. 海洋产几丁质酶菌株的筛选鉴定、基因组分析及几丁质酶酶学性质评价 [D]. 武汉: 武汉轻工大学, 2023. |
| Lu CQ. Screening, identification, genome analysis and enzymatic properties evaluation of chitinase-producing strain from the sea [D]. Wuhan: Wuhan Polytechnic University, 2023. | |
| 13 | 周玉玲, 蒋思婧, 贺妮莎, 等. 微生物几丁质酶研究进展及其在N-乙酰氨基葡萄糖制备中的应用 [J]. 微生物学报, 2021, 61(8): 2192-2204. |
| Zhou YL, Jiang SJ, He NS, et al. Research progress of microbial chitinase and its application in the preparation of N-acetylglucosamine [J]. Acta Microbiol Sin, 2021, 61(8): 2192-2204. | |
| 14 | Poveda J. Glucosinolates profile of Arabidopsis thaliana modified root colonization of Trichoderma species [J]. Biol Contr, 2021, 155: 104522. |
| 15 | Tue NH, Cat Tuong TG, Trang PTH, et al. Cloning the root-specific Asy promoter and genes encoding chitinase 42 kDa of Trichoderma asperellum into the plant expression vector [J]. J App Biol Biotech, 2022: 7-11. |
| 16 | Liu XG, Yu Y, Liu Q, et al. A Na2CO3-responsive chitinase gene from Leymus chinensis improve pathogen resistance and saline-alkali stress tolerance in transgenic tobacco and maize [J]. Front Plant Sci, 2020, 11: 504. |
| 17 | Kabir SR, Rahman MM, Tasnim S, et al. Purification and characterization of a novel chitinase from Trichosanthes dioica seed with antifungal activity [J]. Int J Biol Macromol, 2016, 84: 62-68. |
| 18 | 王琳, 陈雅如, 程湄婕, 等. 微生物几丁质酶研究进展及应用 [J]. 中国生物工程杂志, 2022, 42(12): 101-110. |
| Wang L, Chen YR, Cheng MJ, et al. Research advances in microbial chitinase and its applications [J]. China Biotechnol, 2022, 42(12): 101-110. | |
| 19 | Limón MC, Lora JM, García I, et al. Primary structure and expression pattern of the 33-kDa chitinase gene from the mycoparasitic fungus Trichoderma harzianum [J]. Curr Genet, 1995, 28(5): 478-483. |
| 20 | Wang C, Zeng ZQ, Zhuang WY. Comparative molecular evolution of chitinases in ascomycota with emphasis on mycoparasitism lifestyle [J]. Microb Genom, 2021, 7(9): 000646. |
| 21 | 咸洪泉, 张磊, 李雅华, 等. 特异性顺式作用元件及含该顺式作用元件的启动子和核酸构建体及其应用: CN106754915B [P]. 2019-07-26. |
| Xian HQ, Zhang L, Li YH, et al. Specific cis-acting elements, promoters and nucleic acid constructs containing such cis-acting elements, and their applications: CN106754915B [P]. 2019-07-26. | |
| 22 | Poria V, Rana A, Kumari A, et al. Current perspectives on chitinolytic enzymes and their agro-industrial applications [J]. Biology, 2021, 10(12): 1319. |
| 23 | Jeong GJ, Khan F, Tabassum N, et al. Chitinases as key virulence factors in microbial pathogens: Understanding their role and potential as therapeutic targets [J]. Int J Biol Macromol, 2023, 249: 126021. |
| 24 | Badrhadad A, Nazarian-Firouzabadi F, Ismaili A. Fusion of a chitin-binding domain to an antibacterial peptide to enhance resistance to Fusarium solani in tobacco (Nicotiana tabacum) [J]. 3 Biotech, 2018, 8(9): 391. |
| 25 | Naher L, Yusuf UK, Habib SH, et al. Mycoparasitism activity of Trichoderm harzianum associated with chitinase expression against Ganoderma boninense [J]. Pak J Bot, 2018, 50(3): 1241-1245. |
| 26 | 韩静, 安一博, 季世达, 等. 棘孢木霉Myb27转录因子基因特性分析、原核表达及产物纯化 [J]. 吉林农业大学学报, 2024, 46(1): 49-57. |
| Han J, An YB, Ji SD, et al. Characteristics analysis, prokaryotic expression and product purification of Trichoderma asperellum Myb27 transcription factor [J]. J Jilin Agric Univ, 2024, 46(1): 49-57. | |
| 27 | Ptashne M. How eukaryotic transcriptional activators work [J]. Nature, 1988, 335(6192): 683-689. |
| 28 | Maués DB, Maraschin JC, Duarte DÂ, et al. Overexpression of the transcription factor Azf1 reveals novel regulatory functions and impacts β-glucosidase production in Trichoderma reesei [J]. J Fungi, 2023, 9(12): 1173. |
| 29 | Mu YT, Dong YH, Li XC, et al. JrPHL8-JrWRKY4-JrSTH2L module regulates resistance to Colletotrichum gloeosporioides in walnut [J]. Hortic Res, 2024, 11(7): uhae148. |
| 30 | 王秋月, 段鹏亮, 李海笑, 等. 玉米大斑病菌cDNA文库的构建及转录因子StMR1互作蛋白的筛选 [J]. 生物技术通报, 2024, 40(6): 281-289. |
| Wang QY, Duan PL, Li HX, et al. Construction of cDNA library of Setosphaeria Turcica and screening of transcription factor StMR1 interacting proteins [J]. Biotechnol Bull, 2024, 40(6): 281-289. |
| [1] | YANG Chun, WANG Xiao-qian, WANG Hong-jun, CHAO Yue-hui. Cloning, Subcellular Localization and Expression Analysis of MtZHD4 Gene from Medicago truncatula [J]. Biotechnology Bulletin, 2025, 41(5): 244-254. |
| [2] | HU Ruo-qun, ZENG Jing-jing, LIANG Wan-feng, CAO Jia-yu, HUANG Xiao-wei, LIANG Xiao-ying, QIU Ming-yue, CHEN Ying. Integrated Transcriptome and Metabolome Analysis to Explore the Carotenoid Synthesis and Metabolism Mechanism in Anoectochilus roxburghii under Different Shading Conditions [J]. Biotechnology Bulletin, 2025, 41(5): 231-243. |
| [3] | ZHOU Zhi-guo, FAN Shuang-hu, DENG Chen, FENG Xue. Effects of Exogenous 2,4-Epibrassinolide on Physiological Characteristics of Daucus carota L. Seedlings under Cadmium Stress [J]. Biotechnology Bulletin, 2025, 41(5): 165-174. |
| [4] | PENG Shao-zhi, WANG Deng-ke, ZHANG Xiang, DAI Xiong-ze, XU Hao, ZOU Xue-xiao. Cloning, Expression Characteristics and Functional Verification of the Pepper CaFD1 Gene [J]. Biotechnology Bulletin, 2025, 41(5): 153-164. |
| [5] | LUO Si-fang, ZHANG Zu-ming, XIE Li-fang, GUO Zi-jing, CHEN Zhao-xing, YANG Yue-hua, YAN Xiang, ZHANG Hong-ming. Genome-wide Identification of GATA Gene Family of Jindou Kumquat (Fortunella hindsii) and Their Expression Analysis in Fruit Development [J]. Biotechnology Bulletin, 2025, 41(5): 218-230. |
| [6] | YANG Chao-jie, ZHANG Lan, CHEN Hong, HUANG Juan, SHI Tao-xiong, ZHU Li-wei, CHEN Qing-fu, LI Hong-you, DENG Jiao. Functional Identification of the Transcription Factor Gene FtbHLH3 in Regulating Flavonoid Biosynthesis in Fagopyrum tataricum [J]. Biotechnology Bulletin, 2025, 41(4): 134-144. |
| [7] | WANG Tian-xi, YANG Bing-song, PAN Rong-jun, GAI Wen-xian, LIANG Mei-xia. Identification of the Apple PLATZ Gene Family and Functional Study of the MdPLATZ9 Gene [J]. Biotechnology Bulletin, 2025, 41(4): 176-187. |
| [8] | MA Li-hua, HOU Meng-juan, ZHU Xin-xia. Functional Studies of the Gossypium hirsutumGhNFD4 Gene in Response to Drought in Cotton [J]. Biotechnology Bulletin, 2025, 41(3): 104-111. |
| [9] | SONG Shu-yi, JIANG Kai-xiu, LIU Huan-yan, HUANG Ya-cheng, LIU Lin-ya. Identification of the TCP Gene Family in Actinidia chinensis var. Hongyang and Their Expression Analysis in Fruit [J]. Biotechnology Bulletin, 2025, 41(3): 190-201. |
| [10] | WANG Bin, WANG Yu-kun, XIAO Yan-hui. Comparative Transcriptomic Analysis of Clove Basil (Ocimum gratissimum) Leaves in Response to Cadmium Stress [J]. Biotechnology Bulletin, 2025, 41(3): 255-270. |
| [11] | LIU Jie, WANG Fei, TAO Ting, ZHANG Yu-jing, CHEN Hao-ting, ZHANG Rui-xing, SHI Yu, ZHANG Yi. Overexpression of SlWRKY41 Improves the Tolerance of Tomato Seedlings to Drought [J]. Biotechnology Bulletin, 2025, 41(2): 107-118. |
| [12] | LI Yan-wei, YANG Yan-yan, SUN Ya-ling, HUO Yu-meng, WANG Zhen-bao, LIU Bing-jiang. Regulation Mechanism of Plant Hormones Related to Onion Bulb Enlargement and Development Based on Transcriptome Analysis [J]. Biotechnology Bulletin, 2025, 41(2): 187-201. |
| [13] | QIAN Zheng-yi, WU Shao-fang, CAO Shu-yi, SONG Ya-xin, PAN Xin-feng, LI Zhao-wei, FAN Kai. Identification of the NAC Transcription Factors in Nymphaea colorata and Their Expression Analysis [J]. Biotechnology Bulletin, 2025, 41(2): 234-247. |
| [14] | HUANG Ying, YU Wen-jing, LIU Xue-feng, DIAO Gui-ping. Bioinformatics and Expression Pattern Analysis of Glutathione S-transferase in Populus davidiana × P. bolleana [J]. Biotechnology Bulletin, 2025, 41(2): 248-256. |
| [15] | SHEN Chuan, LI Xia, QIN Jian-feng, DUAN Long-fei, LIU Jia. Screening for WRKY72-interacting Proteins Using a Yeast Two-hybrid Library Derived from Soft Rot Pathogen-induced Amorphophallus konjac [J]. Biotechnology Bulletin, 2025, 41(1): 85-94. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||