








Biotechnology Bulletin ›› 2025, Vol. 41 ›› Issue (7): 205-213.doi: 10.13560/j.cnki.biotech.bull.1985.2025-0069
Previous Articles Next Articles
FU Bo-han( ), MAO Hua, ZHAO Xin-cheng, LU Hong, OU Yong-bin(
), MAO Hua, ZHAO Xin-cheng, LU Hong, OU Yong-bin( ), YAO Yin-an
), YAO Yin-an
Received:2025-01-16
Online:2025-07-26
Published:2025-07-22
Contact:
OU Yong-bin
E-mail:2480198434@qq.com;oyb84@swust.edu.cn
FU Bo-han, MAO Hua, ZHAO Xin-cheng, LU Hong, OU Yong-bin, YAO Yin-an. Cloning of SOS1 Gene Promoters from Poplar and Analysis of Its Response to Salt Stress[J]. Biotechnology Bulletin, 2025, 41(7): 205-213.
| 引物Primer | 序列Sequence (5′-3′) | 功能Function |
|---|---|---|
qRT-PopSOS1-F qRT-PopSOS1-R | ACTTGGCTTCCCATGAACCT AGCAATACCACTGTCACCAA | 检测SOS1基因表达水平 Detection of SOS1 gene expression level |
qRT-PopUBQ-F qRT-PopUBQ-R | CCAAGCCCAAGAAGATCAAGC GCACCGCACTCAGCATTAGG | 内参,校正上样量的差异 Internal reference, correcting for loading variance |
pro-PopSOS1Pro-F pro-PopSOS1Pro-R | GG CC | 克隆杨树SOS1基因启动子 Cloning of the promoters of poplar SOS1 genes |
NPT Ⅱ-F NPT Ⅱ-R | GCTATGACTGGGCACAACAG ATACCGTAAAGCACGAGGAA | 鉴定转基因阳性植株 Checking transgenic positive plants |
Table 1 Primers used in this study
| 引物Primer | 序列Sequence (5′-3′) | 功能Function |
|---|---|---|
qRT-PopSOS1-F qRT-PopSOS1-R | ACTTGGCTTCCCATGAACCT AGCAATACCACTGTCACCAA | 检测SOS1基因表达水平 Detection of SOS1 gene expression level |
qRT-PopUBQ-F qRT-PopUBQ-R | CCAAGCCCAAGAAGATCAAGC GCACCGCACTCAGCATTAGG | 内参,校正上样量的差异 Internal reference, correcting for loading variance |
pro-PopSOS1Pro-F pro-PopSOS1Pro-R | GG CC | 克隆杨树SOS1基因启动子 Cloning of the promoters of poplar SOS1 genes |
NPT Ⅱ-F NPT Ⅱ-R | GCTATGACTGGGCACAACAG ATACCGTAAAGCACGAGGAA | 鉴定转基因阳性植株 Checking transgenic positive plants |
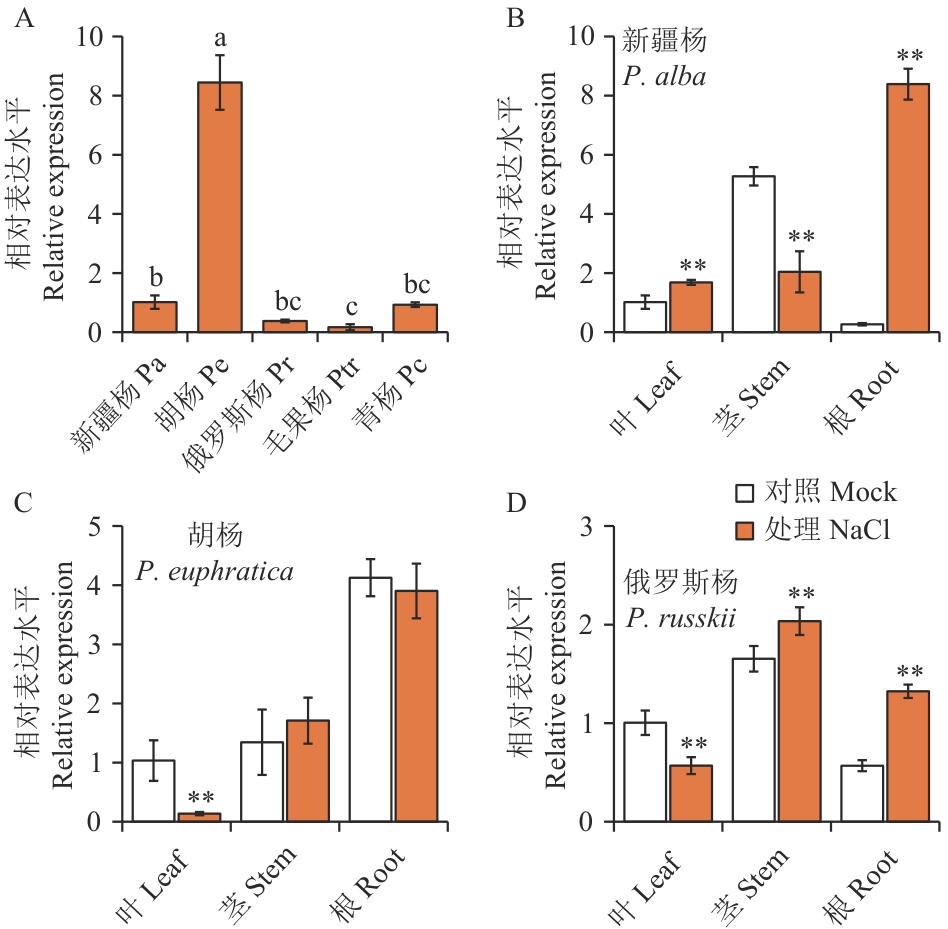
Fig. 1 Relative expressions of SOS1 genes in different poplar species and their responses to salt stressA: The relative expressions of SOS1 in the leaves of P. alba (Pa), P. euphratica (Pe), P. russkii (Pr), P. trichocarpa (Ptr) and P. cathayana (Pc). The relative expression of SOS1 in the leaves of Pa was set to 1. Different normal letters indicate statistical significance among different tree species at P<0.05 level by Duncan's test. B, C, D: The response to salt stress of SOS1 genes in different parts of Pa (B), Pe (C) and Pr (D). Two-month-old greenhouse potted cuttings of Pa or Pr, or six-month-old seedlings of Pe were used as experimental materials for salt stress treatment. In pots with 5 L soils, 200 mmol/L NaCl solution was irrigated every 12 h, 250 mL each time, for a total of 4 times. Samples were collected 48 h after the first irrigation. The same way, 1 L of deionized water was irrigated as the mock. The expressions of SOS1 genes in the leaves of the mock group of each tree species was set to 1. ** indicates statistical significance between the treatment and the mock (t-test, P<0.01)

Fig. 2 Construction of vectors for poplar SOS1 promoters and the identification of transgenic linesA: Electrophoresis detection of the amplified fragment of the poplar SOS1 promoters. B: Cis-acting elements of the PopulusSOS1 gene promoters. C: Double digestion of the promoter vectors. D: Schematic diagrams of the vectors. E: Identification of transgenic plants by PCR. F: GUS histochemical staining of transgenic plantlet leaves. M indicates Trans2K Plus DNA marker (A), TaKaRa DL15000 (C), and Trans15K (E); Pa: P. alba, Pe: P. euphratica, Pr: P. russkii; TSS: transcription start site; ATG: start codon. NPT Ⅱ: Neomycin phosphotransferase Ⅱ; NOS-T: nopaline synthase gene terminator. +: Positive control; -: negative control; CaMV35Spro: Cauliflower mosaic virus 35S promoter; Pa-1, Pa-2, Pa-6, etc. are different transgenic lines

Fig. 3 GUS histochemical staining and quantitative analysis of GUS activity in transgenic poplar under salt stressGUS histochemical staining of leaf discs (A) collected from mature leaves using a punch, transverse section of the 5th internode of the stem (B), and root tips (C). D: GUS activity of different plant parts. Sand-cultured poplars were treated with 1/2 Hoagland solution containing 100 mmol/L NaCl for 7 d before sampling. The mock was cultured with 1/2 Hoagland solution without additional NaCl. Three lines were used for each promoter, and the change pattern was consistent, thus the staining results of only one of them were shown. Scale bars: 1 cm (A) or 1 mm (B and C). * and ** indicate statistical significance by t-test between the treatment and the mock at P<0.05 or P<0.01, respectively
| [1] | Munns R, Tester M. Mechanisms of salinity tolerance [J]. Annu Rev Plant Biol, 2008, 59: 651-681. |
| [2] | 王甜甜, 郝怀庆, 冯雪, 等. 植物HKT蛋白耐盐机制研究进展 [J]. 植物学报, 2018, 53(5): 710-725. |
| Wang TT, Hao HQ, Feng X, et al. Research advances in the function of the high-affinity K+ transporter (HKT) proteins and plant salt tolerance [J]. Chinese Bull Bot, 2018, 53(5): 710-725. | |
| [3] | Navada S, Vadstein O, Gaumet F, et al. Biofilms remember: Osmotic stress priming as a microbial management strategy for improving salinity acclimation in nitrifying biofilms [J]. Water Res, 2020, 176: 115732. |
| [4] | Almeida DM, Margarida Oliveira M, Saibo NJM. Regulation of Na+ and K+ homeostasis in plants: towards improved salt stress tolerance in crop plants [J]. Genet Mol Biol, 2017, 40(1 ): 326-345. |
| [5] | Pan T, Liu MM, Kreslavski VD, et al. Non-stomatal limitation of photosynthesis by soil salinity [J]. Crit Rev Environ Sci Technol, 2021, 51(8): 791-825. |
| [6] | Brett CL, Donowitz M, Rao R. Evolutionary origins of eukaryotic sodium/proton exchangers [J]. Am J Physiol Cell Physiol, 2005, 288(2): C223-C239. |
| [7] | Blumwald E, Aharon GS, Apse MP. Sodium transport in plant cells [J]. Biochim Biophys Acta, 2000, 1465(1/2): 140-151. |
| [8] | Ali A, Petrov V, Yun DJ, et al. Revisiting plant salt tolerance: novel components of the SOS pathway [J]. Trends Plant Sci, 2023, 28(9): 1060-1069. |
| [9] | Shi H, Ishitani M, Kim C, et al. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter [J]. Proc Natl Acad Sci USA, 2000, 97(12): 6896-6901. |
| [10] | 朱业胜, 伍国强, 魏明. 质膜Na+/H+逆向转运蛋白SOS1在植物离子稳态平衡中的作用 [J]. 生物技术通报, 2023, 39(12): 16-32. |
| Zhu YS, Wu GQ, Wei M. Roles of plasma membrane Na+/H+ antiporter SOS1 in maintaining ionic homeostasis of plants [J]. Biotechnol Bull, 2023, 39(12): 16-32. | |
| [11] | Olías R, Eljakaoui Z, Li J, et al. The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs [J]. Plant Cell Environ, 2009, 32(7): 904-916. |
| [12] | Wu SJ, Ding L, Zhu JK. SOS1, a genetic locus essential for salt tolerance and potassium acquisition [J]. Plant Cell, 1996, 8(4): 617-627. |
| [13] | Martínez-Atienza J, Jiang XY, Garciadeblas B, et al. Conservation of the salt overly sensitive pathway in rice [J]. Plant Physiol, 2007, 143(2): 1001-1012. |
| [14] | Xu HX, Jiang XY, Zhan KH, et al. Functional characterization of a wheat plasma membrane Na+/H+ antiporter in yeast [J]. Arch Biochem Biophys, 2008, 473(1): 8-15. |
| [15] | Qiu QS, Guo Y, Dietrich MA, et al. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3 [J]. Proc Natl Acad Sci USA, 2002, 99(12): 8436-8441. |
| [16] | Liu J, Ishitani M, Halfter U, et al. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance [J]. Proc Natl Acad Sci USA, 2000, 97(7): 3730-3734. |
| [17] | Liu J, Zhu JK. A calcium sensor homolog required for plant salt tolerance [J]. Science, 1998, 280(5371): 1943-1945. |
| [18] | Lu KK, Song RF, Guo JX, et al. CycC1;1-WRKY75 complex-mediated transcriptional regulation of SOS1 controls salt stress tolerance in Arabidopsis [J]. Plant Cell, 2023, 35(7): 2570-2591. |
| [19] | Tuskan GA, Difazio S, Jansson S, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) [J]. Science, 2006, 313(5793): 1596-1604. |
| [20] | 谢望, 李天静, 李鑫窈, 等. 胡杨PeNAC121基因启动子的分离鉴定和胁迫应答模式分析 [J]. 植物研究, 2022, 42(2): 234-242. |
| Xie W, Li TJ, Li XY, et al. Identification of PeNAC121 gene promoter and stress response pattern analysis in Populus euphratica [J]. Bull Bot Res, 2022, 42(2): 234-242. | |
| [21] | 高永峰, 杨丰铭, 李琴中, 等. 番茄SlWRKY31基因启动子的克隆与逆境应答模式分析 [J]. 西北植物学报, 2018, 38(12): 2155-2164. |
| Gao YF, Yang FM, Li QZ, et al. Cloning and analysis of stress response pattern of SlWRKY31 gene promoter from tomato [J]. Acta Bot Boreali Occidentalia Sin, 2018, 38(12): 2155-2164. | |
| [22] | Reguera M, Bassil E, Blumwald E. Intracellular NHX-type cation/H+ antiporters in plants [J]. Mol Plant, 2014, 7(2): 261-263. |
| [23] | Zhou HP, Wang CW, Tan TH, et al. Patellin1 negatively modulates salt tolerance by regulating PM Na+/H+ antiport activity and cellular redox homeostasis in Arabidopsis [J]. Plant Cell Physiol, 2018, 59(10): 2165. |
| [24] | Shi HZ, Quintero FJ, Pardo JM, et al. The putative plasma membrane Na(+)/H(+) antiporter SOS1 controls long-distance Na(+) transport in plants [J]. Plant Cell, 2002, 14(2): 465-477. |
| [25] | 黄伦增, 许云泓, 孟凡文, 等. 杨树PtTST3基因启动子克隆及表达活性分析 [J]. 分子植物育种, 2022, 20(17): 5649-5657. |
| Huang LZ, Xu YH, Meng FW, et al. Cloning and expression activity analysis of PtTST3 gene promoter in poplar [J]. Mol Plant Breeding, 2022, 20(17): 5649-5657. | |
| [26] | Guan QM, Wu JM, Yue XL, et al. A nuclear calcium-sensing pathway is critical for gene regulation and salt stress tolerance in Arabidopsis [J]. PLoS Genet, 2013, 9(8): e1003755. |
| [27] | Liscovitch-Brauer N, Montalbano A, Deng JL, et al. Profiling the genetic determinants of chromatin accessibility with scalable single-cell CRISPR screens [J]. Nat Biotechnol, 2021, 39(10): 1270-1277. |
| [28] | Inaba Y, Zhong WQ, Zhang XH, et al. Specificity of expression of the GUS reporter gene (uidA) driven by the tobacco ASA2 promoter in soybean plants and tissue cultures [J]. J Plant Physiol, 2007, 164(7): 824-834. |
| [29] | Maghuly F, Khan MA, Fernandez EB, et al. Stress regulated expression of the GUS-marker gene (uidA) under the control of plant calmodulin and viral 35S promoters in a model fruit tree rootstock: Prunus incisa x Serrula [J]. J Biotechnol, 2008, 135(1): 105-116. |
| [30] | El Mahi H, Pérez-Hormaeche J, De Luca A, et al. A critical role of sodium flux via the plasma membrane Na+/H+ exchanger SOS1 in the salt tolerance of rice [J]. Plant Physiol, 2019, 180(2): 1046-1065. |
| [1] | WANG Fang, QIAO Shuai, SONG Wei, CUI Peng-juan, LIAO An-zhong, TAN Wen-fang, YANG Song-tao. Genome-wide Identification of the IbNRT2 Gene Family and Its Expression in Sweet Potato [J]. Biotechnology Bulletin, 2025, 41(7): 193-204. |
| [2] | ZHANG Ze, YANG Xiu-li, NING Dong-xian. Identification of 4CL Gene Family in Arachis hypogaea L. and Expression Analysis in Response to Drought and Salt Stress [J]. Biotechnology Bulletin, 2025, 41(7): 117-127. |
| [3] | CHENG Shan, WANG Hui-wei, CHEN Chen, ZHU Ya-jing, LI Chun-xin, BIE Hai, WANG Shu-feng, CHEN Xian-gong, ZHANG Xiang-ge. Cloning of MYB Transcription Factor Gene CeMYB154 and Analysis of Salt Tolerance Function in Cyperus esculentus [J]. Biotechnology Bulletin, 2025, 41(6): 218-228. |
| [4] | LI Xiao-huan, CHEN Xiang-yu, TAO Qi-yu, ZHU Ling, TANG Ming, YAO Yin-an, WANG Li-jun. Effects of PtoMYB61 on Lignin Biosynthesis and Salt Tolerance in Populus tomentosa [J]. Biotechnology Bulletin, 2025, 41(6): 284-296. |
| [5] | YANG Chun, WANG Xiao-qian, WANG Hong-jun, CHAO Yue-hui. Cloning, Subcellular Localization and Expression Analysis of MtZHD4 Gene from Medicago truncatula [J]. Biotechnology Bulletin, 2025, 41(5): 244-254. |
| [6] | SONG Hui-yang, SU Bao-jie, LI Jing-hao, MEI Chao, SONG Qian-na, CUI Fu-zhu, FENG Rui-yun. Cloning and Functional Analysis of the StAS2-15 Gene in Potato under Salt Stress [J]. Biotechnology Bulletin, 2025, 41(5): 119-128. |
| [7] | LI Zhi-qiang, LUO Zheng-qian, XU Lin-li, ZHOU Guo-hui, QU Si-yu, LIU En-liang, XU Dong-ting. Identification of the Soybean R2R3-MYB Gene Family Based on the T2T Genome and Their Expression Analysis under Drought and Salt Stress [J]. Biotechnology Bulletin, 2025, 41(5): 141-152. |
| [8] | LIU Tao, WANG Zhi-qi, WU Wen-bo, SHI Wen-ting, WANG Chao-nan, DU Chong, YANG Zhong-min. Identification and Expression Analysis of the GRAM Gene Family in Potato [J]. Biotechnology Bulletin, 2025, 41(4): 145-155. |
| [9] | LIANG Li-cun, WANG Ke-fen, SONG Zu-huan, LIU Meng-ting, LI Jia-yu, LUO Hui-ying, YAO Bin, YANG Hao-meng. Improving the Efficiency of Gene Editing by Optimizing sgRNA in Aspergillus tubingensis [J]. Biotechnology Bulletin, 2025, 41(3): 62-70. |
| [10] | LIN Zi-yi, WU Yi-zhou, YE Fang-xian, ZHU Shu-ying, LIU Yan-min, LIU Su-shuang. Functional Analysis of Soybean GmPM31 Gene Promoter Involvement in Response to High Temperature and Humidity Stress [J]. Biotechnology Bulletin, 2025, 41(3): 90-97. |
| [11] | LI Cai-xia, LI Yi, MU Hong-xiu, LIN Jun-xuan, BAI Long-qiang, SUN Mei-hua, MIAO Yan-xiu. Identification and Bioinformatics Analysis of the bHLH Transcription Factor Family in Cucurbita moschata Duch. [J]. Biotechnology Bulletin, 2025, 41(1): 186-197. |
| [12] | YUAN Liu-jiao, HUANG Wen-lin, CHEN Chong-zhi, LIANG Min, HUANG Zi-qi, CHEN Xue-xue, CHEN Ri-Meng, WANG Li-yun. Effects of Salt Stress on Physiological Characteristics, Ultrastructure and Medicinal Components of Pogostemon cablin Leaves [J]. Biotechnology Bulletin, 2025, 41(1): 230-239. |
| [13] | KOU Bei-sen, CHENG Meng-meng, GUO Xue-qin, GE Bin, LIU Di, LU Hai, LI Hui. Effects of Histone Deacetylase Inhibitor TSA Treatment on the Stem Development of Poplar [J]. Biotechnology Bulletin, 2025, 41(1): 240-251. |
| [14] | WU Shuai, XIN Yan-ni, MAI Chun-hai, MU Xiao-ya, WANG Min, YUE Ai-qin, ZHAO Jin-zhong, WU Shen-jie, DU Wei-jun, WANG Li-xiang. Genome-wide Identification and Stress Response Analysis of Soybean GS Gene Family [J]. Biotechnology Bulletin, 2024, 40(8): 63-73. |
| [15] | WU Shu-ning, SU Yong-ping, LI Dong-xue, BAI Ying-guo, LIU Bo, ZHANG Zhi-wei. Design and Application of a Cumate-inducible Promoter for Corynebacterium glutamicum [J]. Biotechnology Bulletin, 2024, 40(7): 108-116. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||